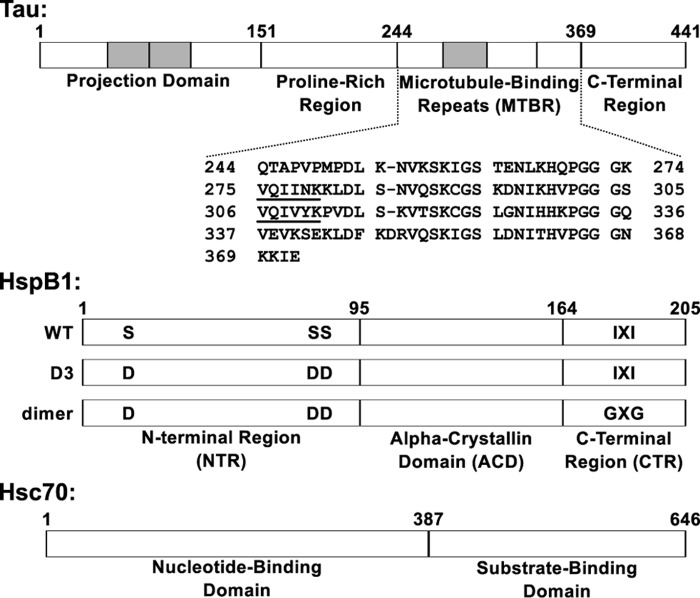
Figure 1.
Domain organization of tau, HspB1, and Hsc70. Top, full-length tau2N4R is natively unstructured and consists of four domains. The tau4RD construct used in this work contains residues 244–372, which include the four microtubule-binding repeats. The sequences 275VQIINK280 and 306VQIVYK311 at the beginning of the second and third repeats drive tau fibril formation. Shaded areas, regions that can be alternatively spliced. Middle, HspB1 contains three domains. The core ACD has a β-sandwich fold and forms a stable dimer. The CTR is intrinsically disordered and contains an IXI motif that interacts with the ACD in the context of large oligomers. The NTR is partially disordered and involved in oligomerization, and it contains three serine residues that are phosphorylated in response to cellular stress. The “D3” and “dimer” constructs, described under “Results,” affect the oligomerization state of HspB1. Bottom, Hsc70 contains two domains: an N-terminal nucleotide binding domain with ATPase activity and a C-terminal substrate binding domain that interacts with client proteins.