Figure 2.
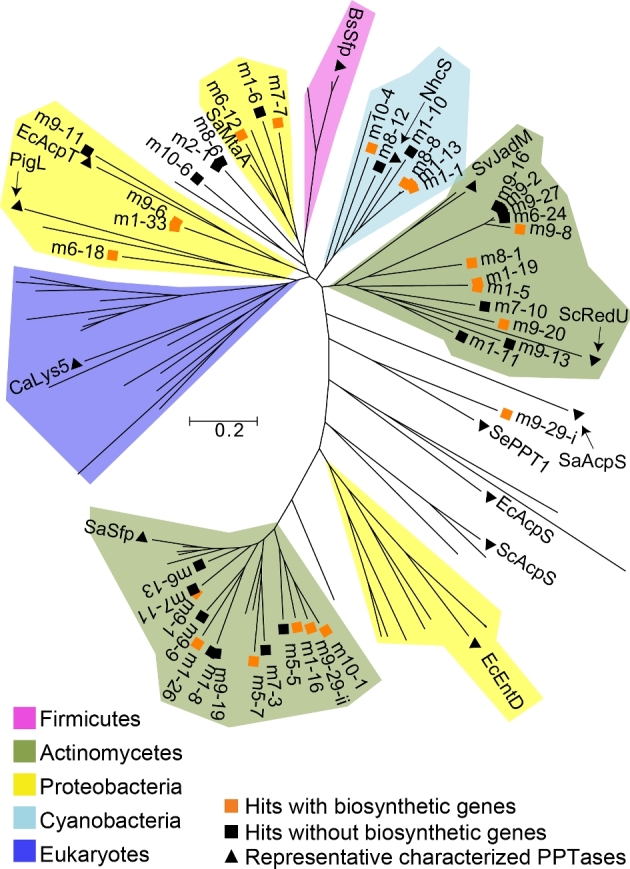
PPTase phylogenetic tree. eDNA-derived PPTases along with ∼60 functionally characterized PPTases of diverse phylogenetic origin (Table S1) were used to construct a phylogenetic tree (Beld et al.2014). Characterized PPTases largely group by phylogenetic origin and therefore clades have been colored using these genes as a guide. Representative functionally characterized PPTases of diverse phylogenetic origin are labeled: S. albus Sfp (SaSfp), E. coli EntD (EcEntD), S. albus AcpS (SaAcpS), S. coelicolor AcpS (ScAcpS), E. coli AcpS (EcAcpS), Saccharopolyspora erythraea PPT1 (SePPT1), S. coelicolor RedU (ScRedU), S. venezuelae JadM (SvJadM), Nodularia spumigena NSOR10 (NhcS), Bacillus subtilis Sfp (BsSfp), S. aurantiaca Sg. a15 MtaA (SaMtaA), E. coli AcpT (EcAcpT), Serratia marcescens PigL (PigL) and Candida albicans Lys5 (CaLys5). The tree was constructed using MEGA (Kumar et al.2016) and the neighbor joining method. A phylogenetic tree where all branches are labeled is provided in Fig. S2.
