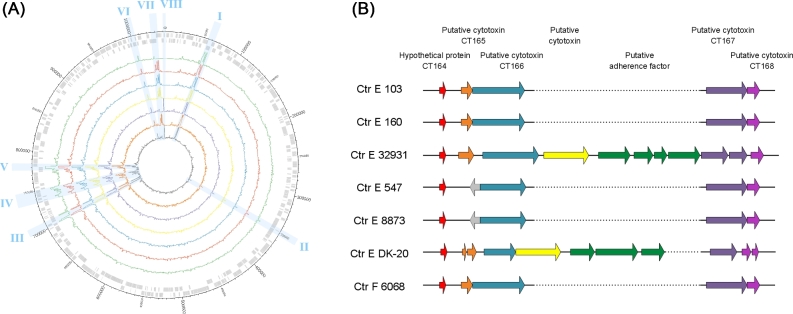
Figure 1.
SNP density and the large insert. (A) This plot shows the SNP densities over the genome sequences of all samples. On the two outermost circles, the genes are plotted in gray, first the forward then the reverse ones. Starting from the outside the genomes are E 103 green, E 160 red, E 32931 blue, E 547 yellow, E 8873 purple, E DK-20 orange and F 6068 gray. The light-blue areas indicate regions with a high SNP density affecting the following proteins. I: Three hypothetical proteins CT_049, CT_050 and CT_051; II: The hypothetical protein CT_310 and the V-type ATP synthase subunit E CT_311; III: Hypothetical protein CT_619 and hypothetical protein CT_622; IV: The intergenic region between formyltetrahydrofolate synthetase CT_649, recombinase RecA CT_650 and a hypothetical protein CT_651; V: major outer membrane protein CT_681; VI: membrane protein CT_852; VII: outer membrane protein PmpE and PmpF, CT_869 and CT_870; VIII: outer membrane protein PmpH CT_872 and hypothetical protein CT_873. (B) Scheme of the region surrounding the large insert plus the predicted annotation of the genes and their homologs in C. trachomatis D/UW-3/CX (CT numbers). Whereas CT_164 is present in all seven samples, the ORF representing CT_165 shows changes in E DK-20 and E 32 931, as well as in E 547 and E 8873, where it is replaced by an ORF on the other strand. CT_166 is also affected by the insert in E 32931 and DK-20. The first ORF of the insert is predicted to be a cytotoxin, followed by three or four putative adherence factors. The insert in E DK-20 is around 900 bp shorter and in E DK-20 and E 32931 either CT_167 or CT_168 is disturbed.