Abstract
Cotton fiber is a specialized unicellular structure useful for the study of cellular differentiation and development. Heat shock proteins (HSPs) have been shown to be involved in various developmental processes. Microarray data analysis of five Gossypium hirsutum genotypes revealed high transcript levels of GhHSP90 and GhHSP70 genes at different stages of fiber development, indicating their importance in the process. Further, we identified 26 and 55 members of HSP90 and HSP70 gene families in G. hirsutum. The treatment of specific inhibitors novobiocin (Nov; HSP90) and pifithrin/2-phenylethynesulfonamide (Pif; HSP70) in in-vitro cultured ovules resulted in a fewer number of fiber initials and retardation in fiber elongation. The molecular chaperone assay using bacterially expressed recombinant GhHSP90-7 and GhHSP70-8 proteins further confirmed the specificity of inhibitors. HSP inhibition disturbs the H2O2 balance that leads to the generation of oxidative stress, which consequently results in autophagy in the epidermal layer of the cotton ovule. Transmission electron microscopy (TEM) of inhibitor-treated ovule also corroborates autophagosome formation along with disrupted mitochondrial cristae. The perturbations in transcript profile of HSP inhibited ovules show differential regulation of different stress and fiber development-related genes and pathways. Altogether, our results indicate that HSP90 and HSP70 families play a crucial role in cotton fiber differentiation and development by maintaining cellular homeostasis.
Introduction
Cotton fiber, one of the longest unicellular cells, provides an ideal platform for studying cellular differentiation, development and cell wall synthesis in the plant system. Cotton fiber development includes four distinct but overlapping stages viz. initiation, elongation, secondary cell wall (SCW) biosynthesis and maturation. Fiber development involves an intricate pattern of transcriptional and translational regulation to facilitate the transition between different stages. An array of genes including transcriptional factors have been reported to play an essential role in fiber initiation and elongation1,2. Apart from this, several metabolic processes such as hormonal pathways2, sugar metabolism
3, secondary metabolites
4, H2O2 balance, etc. are also reported to play a critical role in fiber development. The H2O2 treatment has been shown to induce cotton fiber initials in XinFLM cotton fiber developmental mutant5. Furthermore, treatment of cotton ovules with appropriate concentration of H2O2 prompts fiber elongation via ethylene signaling pathway6,7. However, an increase in the H2O2 concentration beyond optimal level causes oxidative stress, which eventually leads to a decline in the fiber growth8. The imbalance in the levels of H2O2 may cause an adverse effect on the physiology of the organism, by creating stress conditions that may eventually lead to apoptosis9,10. The cell has evolved anti-apoptotic molecules such as HSPs that are induced during the heat stress and delay apoptosis in different cell lines11.
HSPs are a non-identical group of multi-family proteins that are chaperones in nature, primarily predicted to help in the survival of organisms on exposure to stress12. HSPs tend to be an evolutionarily conserved group of proteins throughout the prokaryotes and eukaryotes. These proteins are distributed among five major families viz. HSP100, HSP90, HSP70, HSP60/40, and HSP20 by their molecular weight13. All of these families help in maintaining cellular homeostasis and play distinct non-redundant roles in different developmental processes. Out of these families, HSP90 has been reported to be actively expressed in root and shoot apices14, during embryogenesis and pollen development15 suggesting their role in different developmental processes. HSP90 also contributes to buffering phenotypic variations and developmental stability in Arabidopsis16. HSP90 and HSP70 have been reported to interact with heat shock factors directly and regulate their activity in tomato17. HSP70 also helps in organelle-specific protein sorting18 and directs proteins to ubiquitin-mediated proteasomal degradation pathways19. Down-regulation of HSP70 in Arabidopsis subjects to thermal sensitivity20 and developmental defects21,22. Additionally, HSP70/HSP90 machinery also plays a role in stomatal closure and response to Abscisic acid in Arabidopsis23.
HSPs thus seems to play an essential role in different developmental processes and stress conditions in plants24,25. Studies on several HSP gene families were carried out in various plant species. HSP90 family comprises of 7, 9 and 10 members26,27 whereas, HSP70 family consists of 18, 26 and 20 members in Arabidopsis thaliana, Oryza sativa and Populus trichocarpa, respectively28. Availability of G. hirsutum genome sequence provides the opportunity to explore the HSP gene families in this economically important plant species. Cotton is a field grown plant susceptible to all kind of stresses, there are several reports which point towards the preferential expression of HSPs in different stress conditions, but limited literature is available, indicating towards their involvement in any of its developmental processes29. The commercial availability of HSP specific inhibitors provides a novel platform to explore the importance of HSP proteins in cotton fiber development. Few inhibitors are known for HSP90 and HSP70 but none for HSP100 and HSP20. Recently, butyl 3-[2-(2,4-dichlorophenoxy) acetamido] benzoate has been shown to inhibit HSP60/4030. In plants, HSP90 inhibitor (Geldanamycin) has been shown to decline the root growth in Arabidopsis, suggesting its crucial role in root elongation and thus its suitability for functional studies as HSP90 inhibitor31. Further, HSP90 inhibitor, Nov has been reported to interact with the C-terminal domain of HSP90 that constitutes the dimerization interface and co-chaperone binding domain32. Similarly, HSP70 inhibitor, Pif33 also hinders the co-chaperone and the substrate binding property of HSP70. However, the effect of Pif and Nov in plant system is not analyzed before.
In the present study, we have explored the HSP90 and HSP70 gene families in G. hirsutum and their role in cotton fiber development. We assessed the effect of inhibition of HSP90 and HSP70 in in-vitro ovule culture to study its impact on fiber development. Our results suggest a significant role of HSP70 and HSP90 in maintaining homeostasis during fiber initiation and elongation.
Results
Cotton fiber development concurs with high-level expression of HSPs
All the living organisms are equipped with several classes of structurally unrelated molecular chaperones to ensure proper protein folding during stress condition or rapid development. Analysis of previously published microarray gene expression data34 on six fiber developmental conditions in five genotypes of G. hirsutum, revealed that based on the transcript levels, HSPs are clustered into three distinct clusters (Supplementary Fig. S1). The cluster-II belongs to genes that expressed at high level in almost all the tested developmental stages. The cluster-II consists of different type of HSPs including HSP90 and HSP70 genes, indicating their involvement throughout the fiber development. The transcript level of GhHSP90-7 and GhHSP70-8 belonging to cluster-II was further validated by qRT-PCR, which showed the high transcript level of both these genes especially during initiation and early elongation (Supplementary Fig. S2).
Gene family structure of HSP90 and HSP70 in G. hirsutum
The transcript levels of GhHSP90 and GhHSP70 pointed out their possible role in fiber development. Thus we explored gene family structure of these two chaperons. The HSP90 family in G. hirsutum comprises of total 26 members, 13 each from A and D sub-genomes and contain HSP90 and HATPase_c domain (Table 1). The GhHSP90 family members are distributed on chromosome number (Ch.) 1, 3, 6, 7, 8, 12 and 13 in A sub-genome and on Ch. 1, 2, 3, 6, 7, 8, 12 and 13 in D sub-genome (Table 1). All the homoeologs of GhHSP90 family members are present concurrently on chromosomes of A and D sub-genomes, except for GhHSP90-3A and GhHSP90-3D which are present on Ch. A3 and D2 respectively. Further, GhHSP90-5A.1 and GhHSP90-5A.2, GhHSP90-5D.1 and GhHSP90-5D.2, GhHSP90-7A.1 and GhHSP90-7A.2, GhHSP90-7D.1 and GhHSP90-7D.2 seem to evolve due to independent duplication event. Thus, the presence of four copies each of GhHSP90-5 and GhHSP90-7 in G. hirsutum genome indicates their possible evolutionary importance in growth and development of cotton.
Table 1.
Details of GhHSP90 gene family.
| Gene name | Gene id | Chromosomal position | Protein size (aa) | Molecular Wt. (KDa) | Sub-cellular localization | |
|---|---|---|---|---|---|---|
| A-subgenome | GhHSP90-1A | Gh_A01G0741 | ChrA01: 14221053-14223941 | 699 | 80.036 | Cytoplasmic |
| GhHSP90-2A | Gh_A01G0894 | ChrA01: 21244688-21249882 | 835 | 95.704 | ER | |
| GhHSP90-4A | Gh_A03G0164 | ChrA03: 60064701-60070485 | 698 | 80.155 | Cytoplasmic | |
| GhHSP90-3A | Gh_A03G0935 | ChrA03: 2491019-2494057 | 832 | 94.864 | Cytoplasmic | |
| GhHSP90-5A.1 | Gh_A06G0030 | ChrA06: 223387-227511 | 804 | 91.812 | ER | |
| GhHSP90-5A.2 | Gh_A06G0031 | ChrA06: 229779-233909 | 805 | 92.020 | ER | |
| GhHSP90-6A | Gh_A07G1723 | ChrA07: 70380260-70385380 | 797 | 90.469 | Mitochondrial | |
| GhHSP90-7A.1 | Gh_A08G0219 | ChrA08: 2276489-2279126 | 699 | 80.068 | Cytoplasmic | |
| GhHSP90-7A.2 | Gh_A08G0220 | ChrA08: 2308805-2311733 | 699 | 80.070 | Cytoplasmic | |
| GhHSP90-8A | Gh_A08G0998 | ChrA08: 69729838-69731569 | 318 | 36.430 | Cytoplasmic | |
| GhHSP90-9A | Gh_A12G2300 | ChrA12: 85650149-85653034 | 703 | 80.643 | Cytoplasmic | |
| GhHSP90-10A | Gh_A13G0766 | ChrA13: 32062236-32067412 | 752 | 84.989 | Chloroplastic | |
| GhHSP90-11A | Gh_A13G1098 | ChrA13: 61336732-61339412 | 699 | 80.143 | Cytoplasmic | |
| D-subgenome | GhHSP90-1D | Gh_D01G0761 | ChrD01: 10914595-10917558 | 699 | 80.022 | Cytoplasmic |
| GhHSP90-2D | Gh_D01G0932 | ChrD01: 15653549-15657679 | 809 | 92.490 | ER | |
| GhHSP90-3D | Gh_D02G1319 | ChrD02: 43578662-43584441 | 790 | 90.159 | Cytoplasmic | |
| GhHSP90-4D | Gh_D03G1421 | ChrD03: 42981450-42984339 | 707 | 81.116 | Cytoplasmic | |
| GhHSP90-5D.2 | Gh_D06G2283 | Scaffold4085_D06: 71331-75444 | 804 | 92.044 | ER | |
| GhHSP90-5D.1 | Gh_D06G2284 | Scaffold4085_D06: 64964-69077 | 804 | 92.000 | ER | |
| GhHSP90-6D | Gh_D07G1926 | ChrD07: 47773815-47778936 | 797 | 90.404 | Mitochondrial | |
| GhHSP90-7D.1 | Gh_D08G0299 | ChrD08: 2913021-2915658 | 699 | 80.022 | Cytoplasmic | |
| GhHSP90-7D.2 | Gh_D08G0300 | ChrD08: 2928411-2931340 | 699 | 80.048 | Cytoplasmic | |
| GhHSP90-8D | Gh_D08G1269 | ChrD08: 41600905-41603592 | 704 | 81.051 | Cytoplasmic | |
| GhHSP90-9D | Gh_D12G2436 | ChrD12: 57396182-57399137 | 704 | 80.858 | Cytoplasmic | |
| GhHSP90-10D | Gh_D13G0899 | ChrD13: 18219499-18224694 | 723 | 82.197 | Chloroplastic | |
| GhHSP90-11D | Gh_D13G1363 | ChrD13: 42677111-42679747 | 699 | 80.139 | Cytoplasmic |
ER: Endoplasmic Reticulum.
GhHSP70 family is relatively large consisting of 55 members and characterized by the presence of HSP70 domain. In Arabidopsis, the HSP70 family grouped into HSP70 sub-class and HSP110/SSE sub-class based on their molecular weight28. Similarly in G. hirsutum, out of 55 members, 43 belong to HSP70 sub-class (19 from A and 24 from D sub-genomes) and 12 (6 each from A and D sub-genomes) belong to HSP110/SSE sub-class. The GhHSP70 family members are present on all the chromosomes except Ch. 4 and 7 (Table 2). Like GhHSP90 gene family, all the GhHSP70 genes are located on respective homologous chromosomes in both the sub-genomes, except for GhHSP70-3A and GhHSP70-3D, which is located on A2 and D3 respectively. We failed to detect A sub-genome specific homoeologs of five members namely GhHSP70-5D, GhHSP70-6D, GhHSP70-11D, GhHSP70-16D, and GhHSP70-27D whereas GhHSP70-26D is a partial sequence. The homoeolog of GhHSP70-31A (present on Ch. A10) was identified on scaffold Gh_Sca004937G04 and named as GhHSP70-31D (Table 2).
Table 2.
Details of GhHSP70 gene family.
| Gene name | Gene id | Chromosomal position | Protein size (aa) | Molecular Weight (Da) | Sub-cellular localization | ||
|---|---|---|---|---|---|---|---|
| HSP70 | A-subgenome | GhHSP70-1A | Gh_A01G1923 | ChrA01: 99150074-99153161 | 648 | 71.216 | Cytoplasmic |
| GhHSP70-2A | Gh_A02G0073 | ChrA02: 594987-598554 | 704 | 75.430 | Chloroplastic | ||
| GhHSP70-3A | Gh_A02G0951 | ChrA02: 39564783−39568727 | 666 | 73.381 | ER | ||
| GhHSP70-4A | Gh_A03G0353 | ChrA03: 6375626-6378707 | 648 | 70.824 | Cytoplasmic | ||
| GhHSP70-7A | Gh_A05G0823 | ChrA05: 8267506-8270108 | 650 | 71.113 | Cytoplasmic | ||
| GhHSP70-8A | Gh_A06G1477 | ChrA06: 98009202-98011384 | 648 | 70.997 | Cytoplasmic | ||
| GhHSP70-13A | Gh_A09G1469 | ChrA09: 67930785-67933441 | 646 | 70.869 | Cytoplasmic | ||
| GhHSP70-14A | Gh_A09G2326 | Scaffold2287_A09: 7896-11002 | 718 | 77.299 | Chloroplastic | ||
| GhHSP70-15A | Gh_A09G2245 | Scaffold2279_A09: 7165-9702 | 592 | 64.531 | Cytoplasmic/ PM | ||
| GhHSP70-19A | Gh_A10G1292 | ChrA10: 67260889-67264010 | 706 | 75.703 | Chloroplastic | ||
| GhHSP70-21A | Gh_A11G0171 | ChrA11: 1610300-1613891 | 667 | 73.533 | ER | ||
| GhHSP70-22A | Gh_A11G0174 | ChrA11: 1644547-1647992 | 667 | 73.532 | ER | ||
| GhHSP70-23A | Gh_A11G1883 | ChrA11: 48505796-48509036 | 678 | 72.971 | Mitochondrial | ||
| GhHSP70-24A | Gh_A11G2910 | ChrA11: 93003683-93006152 | 647 | 70.780 | Cytoplasmic | ||
| GhHSP70-25A | Gh_A12G0151 | ChrA12: 2232912-2235970 | 677 | 72.404 | Mitochondrial | ||
| GhHSP70-26A | Gh_A12G0152 | ChrA12: 2269312-2272374 | 681 | 72.814 | Mitochondrial | ||
| GhHSP70-29A | Gh_A13G0895 | ChrA13: 46842882-46846653 | 622 | 69.043 | ER | ||
| GhHSP70-30A | Gh_A13G2046 | ChrA13: 79817836-79819794 | 652 | 71.384 | Cytoplasmic | ||
| GhHSP70-31A | Gh_A10G1940 | ChrA10: 96749888-96752352 | 652 | 71.222 | Cytoplasmic | ||
| D-subgenome | GhHSP70-1D | Gh_D01G2180 | ChrD01: 60746226-60749258 | 648 | 71.204 | Cytoplasmic | |
| GhHSP70-2D | Gh_D02G0088 | ChrD02: 631172-634756 | 704 | 75.441 | Chloroplastic | ||
| GhHSP70-3D | Gh_D03G0811 | ChrD03: 27753247-27756364 | 666 | 73.383 | ER | ||
| GhHSP70-4D | Gh_D03G1221 | ChrD03: 39491745-39494905 | 648 | 70.924 | Cytoplasmic | ||
| GhHSP70-5D | Gh_D03G1225 | ChrD03: 39573593-39578627 | 584 | 65.563 | Cytoplasmic | ||
| GhHSP70-6D | Gh_D03G1549 | ChrD03: 44705138-44707084 | 648 | 70.806 | Cytoplasmic | ||
| GhHSP70-7D | Gh_D05G0943 | ChrD05: 7907708-7910317 | 650 | 71.174 | Cytoplasmic | ||
| GhHSP70-8D | Gh_D06G1814 | ChrD06: 58203414-58205599 | 648 | 71.011 | Cytoplasmic | ||
| GhHSP70-11D | Gh_D08G1192 | ChrD08: 38372483-38375636 | 666 | 73.252 | ER | ||
| GhHSP70-13D | Gh_D09G1479 | ChrD09: 42364418-42367075 | 646 | 70.871 | Cytoplasmic | ||
| GhHSP70-14D | Gh_D09G2036 | ChrD09: 47728996-47732064 | 735 | 79.022 | Chloroplastic | ||
| GhHSP70-15D | Gh_D09G2082 | ChrD09: 48164612-48167151 | 592 | 64.721 | Cytoplasmic/ PM | ||
| GhHSP70-16D | Gh_D09G2084 | ChrD09: 48173223-48174926 | 567 | 62.652 | Cytoplasmic/ PM | ||
| GhHSP70-19D | Gh_D10G1189 | ChrD10: 20558563-20561664 | 706 | 75.708 | Chloroplastic | ||
| GhHSP70-21D | Gh_D11G0181 | ChrD11: 1618339-1621899 | 667 | 73.532 | ER | ||
| GhHSP70-22D | Gh_D11G0184 | ChrD11: 1641418-1644817 | 667 | 73.451 | ER | ||
| GhHSP70-23D | Gh_D11G2087 | ChrD11: 29998341-30002431 | 682 | 73.337 | Mitochondrial | ||
| GhHSP70-24D | Gh_D11G3296 | ChrD11: 65856743-65859212 | 647 | 70.983 | Cytoplasmic | ||
| GhHSP70-25D | Gh_D12G0164 | ChrD12: 2116044-2119093 | 677 | 72.406 | Mitochondrial | ||
| GhHSP70-26D | Gh_D12G0165 | ChrD12: 2130556-2132570 | 354 | 37.57 | Mitochondrial | ||
| GhHSP70-27D | Gh_D12G1067 | ChrD12: 36783328-36784989 | 553 | 60.936 | Cytoplasmic | ||
| GhHSP70-29D | Gh_D13G1136 | ChrD13: 33658463-33661024 | 657 | 72.809 | ER | ||
| GhHSP70-30D | Gh_D13G2447 | ChrD13: 60389215-60391173 | 652 | 71.384 | Cytoplasmic | ||
| GhHSP70-31D | Gh_Sca004937G04 | Scaffold4937: 28658-31068 | 652 | 71.307 | Cytoplasmic | ||
| HSP110/SSE | A-subgenome | GhHSP70-9A | Gh_A06G1513 | ChrA06: 98810230-98817100 | 911 | 101.51 | Nuclear/ ER |
| GhHSP70-10A | Gh_A08G2507 | Scaffold2268_A08: 181657-184884 | 757 | 84.780 | Nuclear | ||
| GhHSP70-12A | Gh_A09G0792 | ChrA09: 54719230-54722953 | 856 | 94.427 | Cytoplasmic | ||
| GhHSP70-17A | Gh_A10G0130 | ChrA10: 1084260-1088698 | 774 | 86.965 | Nuclear | ||
| GhHSP70-18A | Gh_A10G0712 | ChrA10: 12129630-12133303 | 855 | 94.337 | Cytoplasmic | ||
| GhHSP70-20A | Gh_A10G1717 | ChrA10: 91002111-91010963 | 879 | 98.502 | ER/ Nuclear | ||
| D-subgenome | GhHSP70-9D | Gh_D06G2388 | Scaffold4164_D06: 47843-55469 | 913 | 101.573 | Nuclear/ ER | |
| GhHSP70-10D | Gh_D08G0244 | ChrD08: 2325136-2328250 | 757 | 84.872 | Nuclear | ||
| GhHSP70-12D | Gh_D09G0795 | ChrD09: 32510122-32513840 | 856 | 94.599 | Cytoplasmic | ||
| GhHSP70-17D | Gh_D10G0137 | ChrD10: 1086860-1091313 | 774 | 86.814 | Nuclear | ||
| GhHSP70-18D | Gh_D10G0674 | ChrD10: 7564910-7568532 | 855 | 94.195 | Cytoplasmic | ||
| GhHSP70-20D | Gh_D10G1993 | ChrD10: 55383231-55392087 | 879 | 98.686 | ER/ Nuclear |
ER: Endoplasmic Reticulum.
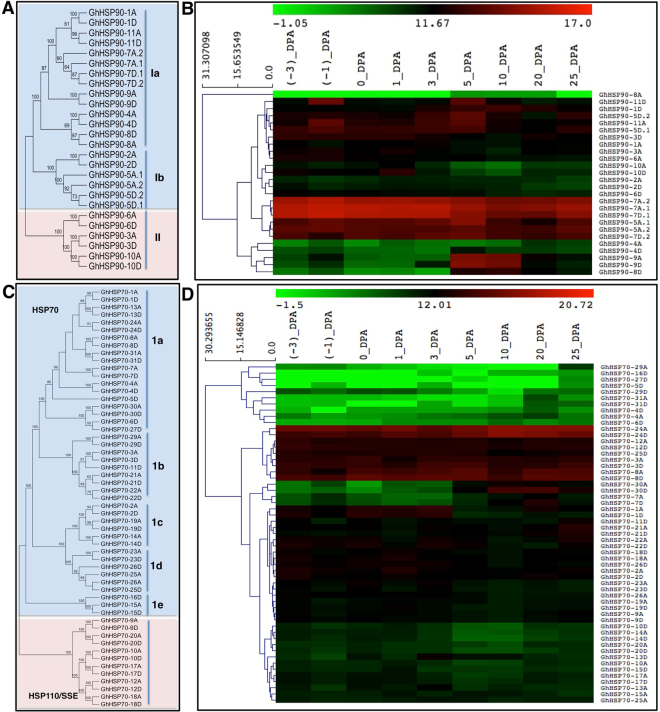
Sub-cellular localization prediction indicated that members of both the families were localized either in the cytoplasm or other sub-cellular organelles. Of all the members of GhHSP90 and GhHSP70 sub-class, 16 and 23 members were cytoplasmic. Other members were found to localize either in the ER (six and nine members respectively), chloroplast (two and five members respectively) or in the mitochondria (two and six members respectively) (Tables 1 and 2). Similarly, the members of HSP110/SSE sub-class were also predicted to localize in the cytoplasm (four members), nucleus (four members), ER/nucleus (two members) or the nucleus/ER (two members) (Table 2). The phylogenetic tree showed GhHSP90 family members grouped mainly into two groups with members of each branch having similar sub-cellular localization. The members of the group I were further sub-categorized into Ia (members localized in the cytoplasm) and Ib (members localized in the ER), whereas group II members were predicted to localize in the chloroplast or mitochondria (Fig. 1A). In the case of GhHSP70, the phylogenetic tree bifurcated the two sub-classes on different branches, which further formed groups based on their sub-cellular localization. The HSP70 subclass grouped into Ia, Ib, Ic, Id, and Ie that were predicted to localize into the cytoplasm, ER, chloroplast, mitochondria, and cyto/PM, respectively (Fig. 1C). Whereas, the HSP110/SSE members formed altogether a different cluster II (Fig. 1C).
Figure 1.
Phylogenetic and transcript level analysis of HSP90 and HSP70 members in G. hirsutum. (A) Phylogenetic tree of GhHSP90 members. (B) Heat map showing transcript levels of GhHSP90 members at different stages of fiber development. (C) Phylogenetic tree of GhHSP70 members. (D) Heat map showing transcript levels of GhHSP70 members at different stages of fiber development.
To probe further into the possible role of GhHSP90 and GhHSP70 genes during fiber development, we investigated their transcript levels. Among GhHSP90 members, GhHSP90-7 and GhHSP90-5A showed higher transcript levels in all the stages of fiber development, suggesting their importance throughout the fiber development. GhHSP90-2 and GhHSP90-6D showed medium transcript levels in all the stages whereas, GhHSP90-4, GhHSP90-9, and GhHSP90-8D showed significant transcript levels in the later stages of development. GhHSP90-10 was found to express significantly in early stages, and GhHSP90-8A has poor transcript levels in all the stages of development (Fig. 1B). Similarly, the members of GhHSP70 were clustered into genes with low transcript levels in all the stages (GhHSP70-29, GhHSP70-16D, GhHSP70-27D, GhHSP70-5D, GhHSP70-31, GhHSP70-4 and GhHSP70-6D), genes with high transcript levels in all the stages (GhHSP70-24, GhHSP70-12, GhHSP70-25D, GhHSP70-3, and GhHSP70-8) and genes with intermediate transcript levels in all the stages (GhHSP70-10, GhHSP70-14, GhHSP70-20, GhHSP70-13, GhHSP70-15, GhHSP70-17, GhHSP70-25A, GhHSP70-23, GhHSP70-26A, GhHSP70-19 and GhHSP70-9) (Fig. 1D). While GhHSP70-30 and GhHSP70-7 showed significant transcript level in the later stages, GhHSP70-1, GhHSP70-22, GhHSP70-18, GhHSP70-26D, GhHSP70-2, GhHSP70-11D and GhHSP70-21 showed higher transcript levels in early stages of fiber development, suggesting their importance in their corresponding stages (Fig. 1D).
HSP90 and HSP70 activities are essential for the appropriate development of cotton fiber
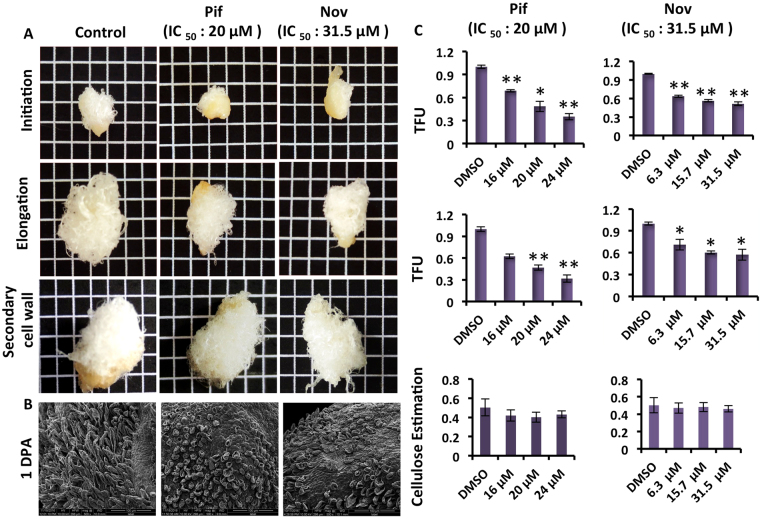
Nov and Pif are reported inhibitors for HSP9032 and HSP7033 classes of proteins, respectively. We assessed the role of HSP90 and HSP70 in cotton fiber development by treating developing cotton ovules with Nov and Pif respectively in in-vitro ovule culture (Fig. 2A). The varying concentrations of Nov and Pif were used to determine the IC50 value (Supplementary Fig. S3). Both the inhibitors showed significant inhibition of fiber development with increasing concentrations as also indicated by decreasing total fiber unit (TFU) (Fig. 2C). This result was further confirmed by the decline in fiber growth observed in scanning electron microscopy (SEM) of inhibitor-treated ovules (Fig. 2B). The inhibition of the fiber development was more pronounced when inhibitors were added either at initiation or at the elongation stage for both Nov and Pif (Fig. 2A). The IC50 for Nov for both the initiation and elongation stages was 31.5 µM, while that of Pif for both stages was 20 µM (*p-value ≤ 0.05, **p-value ≤ 0.01). We did not observe any significant inhibition by either Nov or Pif when treated at SCW biosynthesis stage; this is also indicated by no change in cellulose content (Fig. 2A and C). Thus, our results showed that appropriate chaperonic activities of HSP90 and HSP70 are crucial for optimal fiber development at initiation and elongation stages.
Figure 2.
Effect of different HSP inhibitors on in-vitro ovule culture at different stages of cotton fiber development. (A) Photographs of in-vitro cultured ovules in initiation, elongation and SCW stage showing the effect of Pif (HSP70) and Nov (HSP90) inhibitors. (B) SEM of control and treated ovules at 1 DPA shows clear decline in fiber growth. (C) Quantitative estimation of fiber growth in initiation, elongation and SCW stage at different concentration of Pif and Nov inhibitors (X-axis and Y-axis designates O.D. and concentration of inhibitors, respectively). The asterisks represent statistical significance between three biological and three technical replicates (*p-value ≤ 0.05, **p-value ≤ 0.01).
Nov and Pif are inhibitors of cotton HSP90 and HSP70
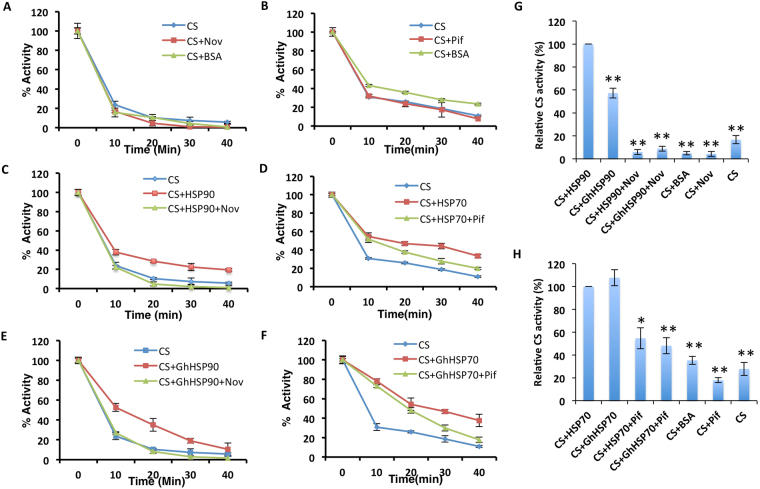
The specificity of inhibition of fiber development by Nov and Pif was evaluated using Citrate synthase (CS) assay35. CS is a substrate for both HSP90 and HSP70, and it is a thermally unstable protein that makes it suitable for analyzing chaperone activities35,36. Initially, the assay was standardized using human CS, HSP90 and HSP70 obtained from Sigma, USA (Fig. 3A–D). The Nov/Pif (Fig. 3A and B) alone do not affect the activities of CS. However, the inclusion of human HSP90 (Fig. 3C) and HSP70 (Fig. 3D) in the reactions showed significant chaperonic thermos-protection as expected. But, BSA at an equivalent concentration does not show thermos-protection on CS (Fig. 3A and B). Further, the addition of Nov to the reaction containing CS and human HSP90 resulted in the loss of thermos-protection by HSP90 (Fig. 3C). Similarly, Pif also inhibits thermos-protection by HSP70 (Fig. 3D). Thus, our results validate previously reported thermos-protection activities of HSP90 and HSP70 on CS35 and the specificity of inhibition of these interactions by Nov and Pif respectively. Next, we replaced the human HSPs from the CS thermos-protection assay with bacterially expressed partially purified recombinant GhHSP90-7 and GhHSP70-8 proteins (Supplementary Fig. S4). We observed, similar to human HSP90 and HSP70, GhHSP90-7 and GhHSP70-8 also showed significant thermos-protection to CS in our assay (Fig. 3E and F). Further, the addition of Nov and Pif completely inhibited the thermos-protection activity of GhHSP90-7 and GhHSP70-8, respectively (Fig. 3E and F). Thus, our results showed that inhibition of fiber development observed in in-vitro condition by Nov and Pif was indeed due to their inhibitory effect on chaperonic activities of GhHSP90 and GhHSP70, respectively.
Figure 3.
Chaperone assay of GhHSP proteins. (A) Effect of control protein BSA and inhibitor Nov on activity of Citrate synthase (CS) (B) Effect of control protein BSA and Pif on activity of CS. (C) Effect of Human HSP90 on activity of CS, with or without Nov. (D) Effect of Human HSP70 on activity of CS, with or without Pif. (E) Effect of GhHSP90 on activity of CS, with or without Nov. (F) Effect of GhHSP70 on activity of CS, with or without Pif. (G) The mean CS activity at the end point of A, C and E relative to CS-thermosprotection in the presence of HSP90 protein. (H) The mean CS activity at the end point of B, D and F relative to CS-thermosprotection in the presence of HSP70 protein. The asterisks represent statistical significance between two independent experiments (*p-value ≤ 0.05, **p-value ≤ 0.01).
Inhibition of HSP90 and HSP70 activity leads to ROS imbalance and autophagy in developing fibers
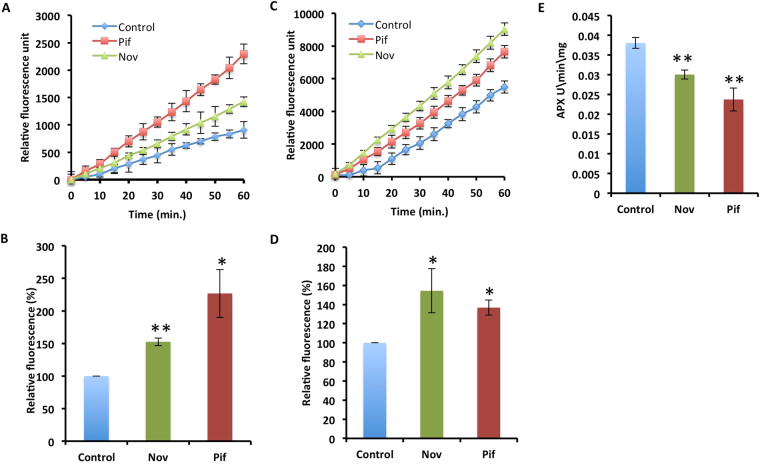
The HSPs are involved in cellular homeostasis during stress and rapid growth. Thus, any imbalance in HSPs like inhibition of HSP activity by inhibitors impaired cellular homeostasis and therefore resulted in ROS imbalance. Fiber development also requires finely tune ROS response5. Thus any imbalance in ROS may lead to improper fiber development. Hence, we examined ROS response in Nov or Pif treated ovules by estimating the H2O2, superoxide, and ascorbate peroxidase (APX) activity7,37,38. The Nov and Pif treated ovules at 0, and 6 Days post anthesis (DPA) in in-vitro condition showed a significant increase in H2O2 and superoxide levels (Fig. 4A,C, and Supplementary Fig. S5). The results suggested that the inhibition of HSPs leads to significantly higher accumulation of H2O2 and superoxide radicals. Further, as expected treatment of Nov or Pif also leads to significant decrease in the APX activity at 0 DPA, which correlates, well with higher H2O2 level (Fig. 4E). Thus, results indicate that treatment of developing fibers with Nov and Pif leads to an imbalance in ROS.
Figure 4.
Biochemical alterations due to HSP inhibitor treatment in in-vitro cultured ovules. (A) Relative H2O2 estimation in Nov and Pif treated ovules at 0 DPA. (B) The mean fluorescence at the end point of Nov and Pif treated ovules as compared to control ovules at 0 DPA. The asterisks represent statistical significance between three biological and two technical replicates (*p-value ≤ 0.05, **p-value ≤ 0.01). (C) Relative H2O2 estimation in Nov and Pif treated ovules at 6 DPA (D) The mean fluorescence at the end point of Nov and Pif treated ovules as compared to control ovules at 6 DPA. The asterisks represent statistical significance between three biological and two technical replicates (*p-value ≤ 0.05, **p-value ≤ 0.01). (E) Quantitative estimation of Ascorbate peroxidase (APX) activity in Nov and Pif treated ovules at 0 DPA (**p-value ≤ 0.01).
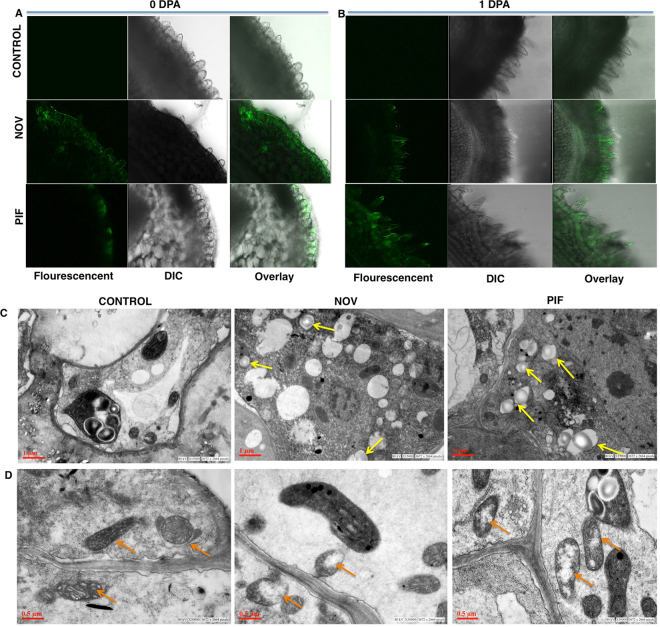
The higher accumulation of H2O2 and inhibition of HSP has shown to destine the cells to autophagy-mediated cell death39,40. Thus, we examined the potential induction of autophagy in Nov and Pif treated fibers at 0 DPA and 1 DPA by staining them with monodansylcadaverine (MDC). The committed fiber cells (0 DPA) and the protruding fibers (1 DPA) showed significant fluorescence in the Nov and Pif treated ovules indicating a higher level of autophagosomes, whereas no significant fluorescence was observed in control ovules treated with DMSO (Fig. 5A and B). Thus, our results revealed that higher accumulation of H2O2 due to inhibition of HSP activities lead to significant induction of autophagy in the fibers treated with Nov and Pif.
Figure 5.
Induction of autophagy in Nov and Pif treated cotton ovules. Monodansylcadaverin (MDC) staining shows the presence of autophagosomes in cotton ovules treated with inhibitors Nov and Pif at (A) 0 DPA (B) 1 DPA (C) TEM of Nov and Pif treated ovules show the presence of autophagosomes (yellow arrow) at 1 DPA (D) TEM of Nov and Pif treated ovules show the presence of disrupted mitochondrial cristae (orange arrow) in treated ovule at 1 DPA.
The TEM analysis of 1 DPA fibers of control or Nov/Pif treated fiber cells further confirmed the results (Fig. 5C). The TEM revealed that Nov and Pif treated fiber cells showed a significantly higher number of refractive autophagosomes, which are altogether absent from the un-treated fiber cells. The previous report also suggested that autophagy caused due to oxidative stress, targets ROS production sites, such as mitochondria41. In TEM images we observed that Nov and Pif treated cells showed abnormal mitochondrion with disorganized cristae while that in control was well formed (Fig. 5D). Thus, results confirm that HSP inhibition causes induction of oxidative stress in the ovule that ultimately leads to autophagy.
HSP90 and HSP70 inhibition result in modulation of the transcriptome during cotton fiber development
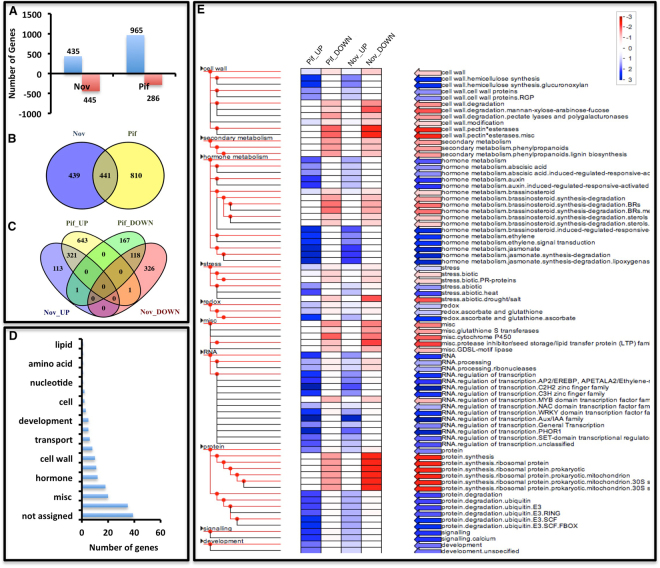
Cotton fiber development is a complex process as it involves a suite of transcription factors and regulators1,2. Application of HSP inhibitors hinders the fiber growth in the developing ovules, which might have accompanied by the pronounced alteration in transcription in developing fibers. The transcriptome sequencing of Nov/Pif treated and control ovules at 6 DPA was carried out (Supplementary Table S2). The quality filtered reads were mapped on G. hirsutum reference genome and identified differentially expressed genes (DEGs). Nov treatment leads to up-regulation of 435 genes and down-regulation of 445 genes (total 880 DEGs) as compared to control (Fig. 6A), whereas, Pif treatment results in a total of 1251 DEGs out of which 965 were up-regulated and 286 were down-regulated in 6 DPA ovules (Fig. 6A). The total of 441 DEGs was found to be common between DEGs of Nov and Pif (Fig. 6B) of which 321 DEGs were up-regulated and 118 DEGs were down-regulated in both the cases (Fig. 6C). The percentage of A or D specific expressed genes in either of the inhibitors treated samples remain almost same (Supplementary Table S3). We analyzed common DEGs to identify pathways influenced by inhibition of HSP90 and HSP70 during fiber development. It was interesting to note that the DEGs belong to up- and down-class for both the inhibitors were strikingly similar (Fig. 6C). The significant characteristic metabolic bins assigned to DEGs in MapMan analysis are RNA, Protein, hormonal metabolism, signaling, cell wall and stress (Fig. 6D). The pathways and genes that were up-regulated due to inhibition of both HSPs belongs to ABA metabolism, auxin metabolism, ethylene metabolism, jasmonate metabolism, abiotic stress, calcium signaling, GABA amino acid metabolism, nucleotide salvage pathways, vacuole protein targeting, kinases, protein degradation, signaling, miscellaneous pathways like UDP glucosyl and glucoronyl transferases, oxidases, invertase, AP2/EREBP, NAC, WRKY, AUX/IAA, PHOR1, potassium transporter, armadillo/beta-catenin repeat protein etc. (Fig. 6E, Supplementary Fig. S6). The pathways and genes that were down-regulated include mitochondrial electron transport, cell wall, lignin biosynthesis, brassinosteroid biosynthesis, lipid metabolism, cytochrome P450, GDSL-motif lipase, MYB40, GST, LTP, protein synthesis, intrinsic protein transporters, etc. (Fig. 6E, Supplementary Fig. S6). These DEGs includes several genes and pathway that were reported to be involved in fiber development such as mitochondrial electron transport, cell wall, phenylpropanoid pathways, brassinosteroid biosynthesis, cytochrome P450, GDSL-motif lipase and MYB transcription factors (Fig. 6E)42–45. Besides, HSP inhibition resulted in up-regulation of several stress-related pathways; these include ABA metabolism, auxin metabolism, ethylene metabolism, jasmonate metabolism, abiotic stress, Calcium, AP2/EREBP, NAC, WRKY, AUX/IAA, etc. (Fig. 6E)46–49. The transcriptome data was further validated using qRT-PCR of seven each of commonly up-regulated (WRKY53, ABA-responsive gene, BRH1, NAM, C2H2-Zn finger, Glycoside hydrolase, and NDR1) and down-regulated genes (Ribosomal protein, WRKY29, GDSL-Lipase, EXP8, UGT72E1, MYB40, and SQE) that were identified using RNAseq (Supplementary Table S4). GbUbiQ1 and Histone3 were used as internal control genes to normalize the real-time expression values (Supplementary Fig. S7). Several genes that were selected for qRT-PCR validation are implicated earlier for their role in the fiber development. The qRT-PCR analysis showed that all the selected genes showed transcript level similar to that observed in RNAseq, thus validating their expression pattern (Supplementary Fig. S7).
Figure 6.
Transcriptomic alterations in HSP90 and HSP70 inhibitor treated in-vitro cultured cotton ovules. (A) Differentially regulated genes in Nov and Pif treated ovules at 6 DPA. (B) Venny diagram showing common genes getting differentially expressed in both Nov and Pif treated ovules. (C) Venny diagram showing regulation status of all the DEGs in both the treatments. (D) Bar chart showing distribution of common DEGs in different biological processes. (E) Pageman showing major pathways regulated by Nov and Pif inhibition in elongation stage.
Discussion
Plants are sessile; therefore, the importance of HSPs in combating stress conditions escalates. HSPs have also been reported to play a crucial role in plant growth and development. Suppression of HSP90 activity leads to developmental defects in Arabidopsis50, suggesting their essential role in plant development. Likewise, HSP70 is known to bind nascent polypeptides helping them to fold, prevent aggregation and keep them in an import-competent state51, also HSP70 has been reported to induce during microspore differentiation in Capsicum52. Several HSP genes were down-regulated at both RNA and protein levels in ligon lintless-1 (Li1) mutant of G. hirsutum, suggesting their involvement in fiber development29,53. Cotton fiber development is a complex process, and differential expression of several members of HSPs in the microarray of different fiber developmental stages indicates their involvement in the process (Supplementary Fig. S1). The consistent higher transcript level of HSP90 and HSP70 genes throughout various stages in all the five superior and inferior fiber quality genotypes of G. hirsutum suggests their pivotal importance in the development of cotton fiber (Supplementary Fig. S1). Further, the real-time expression analysis of these genes in one of the superior fiber quality genotype of G. hirsutum corroborate their role especially in early stages of fiber development, i.e., in fiber initiation and elongation (Supplementary Fig. S2).
HSPs are multigene protein families; different members of HSP gene families might play various roles depending on their pattern of transcript level, substrate specificity, and localization. HSP family members showed differential transcript levels during fiber development (Supplementary Fig. S1). We focused only on HSP90 and HSP70 family since they are predominantly expressed during all the stages of fiber development (Supplementary Fig. S1) in G. hirsutum. Both the HSP90 and HSP70, gene families, remain highly conserved throughout the kingdom, probably due to their involvement in several fundamental biological processes and defense response. The genome-wide analysis shows the occurrence of 26 and 55 members of GhHSP90 and GhHSP70 gene families in G. hirsutum, respectively (Tables 1 and 2). The number of these family members are significantly higher in comparison to the other plant species, probably due to tetraploid nature of G. hirsutum, but paleopolyploid soybean has yet higher, 61 GmHSP70 members54. The duplication of GhHSP90-5 and GhHSP90-7 seems to be evolutionarily relevant, as these genes were also duplicated in the G. arboreum (A) and G. raimondii (D) progenitors and have transferred to G. hirsutum during its speciation. The phylogenetic analysis shows the distribution of GhHSP90 members into two major groups by their cellular localization (Fig. 1A). The members probably have conserved sub-cellular localization due to their specific functions55. GhHSP70 members present into two major groups in the phylogenetic tree, the higher molecular weight members, HSP110/SSE members form one group, and the lower molecular weight members form the other group, probably due to their sub-cellular localization and functional diversity (Fig. 1C). Further, the transcript abundance analysis of GhHSP90 and GhHSP70 genes in different stages of fiber development shows the constitutive as well as stage-specific transcript level suggesting the specific and non-overlapping importance of these members. Most of the HSP90 and HSP70 homeologs show similar transcript levels and clustering (Fig. 1B and D).
HSPs are chaperone proteins that help in maintaining the protein homeostasis56. Cotton fiber development requires a repertoire of genes and proteins expressing during different development stages, the maintenance of cellular homeostasis during the process is a must and may require HSP proteins. Application of HSP90 inhibitor (Radicicol) on developing fibers in-vitro hinders fiber elongation, but the study was limited to brief phenotypic observations29. Several HSP inhibitors have been extensively studied and characterized in the animal system32,33. Application of HSP inhibitors in in-vitro ovule culture can prove to be an efficient system for exploring their role in fiber development. We studied the inhibitory activity of two of the previously reported inhibitors of HSP90 and HSP70, i.e., Nov and Pif, respectively in the plant system for the first time. The decline in fiber growth on the application of HSP inhibitors further strengthen the importance of HSP90 and HSP70 activity in both initiation as well as in elongation stage of fiber development. The phenotypic and biochemical parameters suggest varying concentration of Nov and Pif showed pronounced inhibition of fiber development (Fig. 2A and C). The SEM also shows the significant decline in fiber growth (Fig. 2B). However, no significant alteration observed during secondary cell wall deposition stage (Fig. 2C) indicating that HSPs are essential during rapid growth at fiber initiation and elongation.
Mainly the HSP inhibitors for animal HSPs are biochemically characterized till date. However, these HSP inhibitors can potentially inhibit plant derived HSPs, due to their evolutionarily conserved overall domain structure32,33. The well-established protocol on thermo-stability of CS in presence or absence of Nov and Pif was used to evaluate their specificity in inhibiting chaperonic activities of GhHSP70 and GhHSP90. Plant-derived GhHSP70 and GhHSP90 showed comparable thermos-protection to the human CS confirming their evolutionary conservation of substrate preference (Fig. 3). The thermos-protecting chaperonic activity of GhHSP70 and GhHSP90 inhibited efficiently by Nov and Pif (Fig. 3E and F), pitching on the importance of these molecules for functional studies on HSPs. Further, the mere addition of Nov or Pif to the reactions do not showed any effect on the activity of CS, indicating that the inhibition by Nov and Pif are specific (Fig. 3A and B). The results thus confirmed that inhibition of fiber development by treatment of Nov/Pif was actually due to the inhibition of chaperonic activities of HSP70/90.
The increased levels of H2O2 lead to oxidative stress8. Also, H2O2 balance is crucial in growth and development, including cotton fiber development5. In agreement with the previous studies, we also observed a rise in the H2O2 levels due to HSP inhibition during cotton fiber development57. The imbalance in H2O2 levels leads to inhibition of fiber development in both initiation and elongation stage (Fig. 4A and C). The optimal concentration of H2O2 plays a vital role to destine the cotton ovule epidermal cell to differentiate into a fiber5. The outburst of H2O2 accompanies the transition from initiation to elongation5,6. In our study inhibition of HSP90/70 by Nov and Pif might have resulted in the alteration in cellular homeostasis, which leads to oxidative burst (higher levels of H2O2 and superoxide radicals) and a decline in fiber growth. The expression pattern of HSP90 and HSP70 genes might correlate with H2O2 levels in fiber cells. The lintless-fuzzless mutants show low transcript levels of HSP90 and HSP70 genes42 and also undergo fiber initiation when treated with appropriate concentrations of H2O25.
The higher H2O2 levels have been shown to be detrimental to growth and development in plants9. Antioxidant enzymes, like APX, maintain the balance of H2O2. As expected, lower APX activity leads to higher H2O2 levels in Nov and Pif treated ovules (Fig. 4E), clearly indicating that Nov and Pif treatment leads to oxidative stress like condition in developing fibers. The optimal APX activity is needed for proper development of fibers since lintless-fuzzless mutants showed reduced APX activity7. However, significantly higher levels of oxidative radicals (H2O2 and superoxide) and declined APX activity may have destined fiber cells to death since a rise in the H2O2 signals the cell toward apoptotic pathways, eventually leading to cell death58. We show the accumulation of significantly higher level of autophagosome in both Nov and Pif treated fiber cells (Fig. 5). The autophagy induced due to oxidative stress is accompanied by the destruction of ROS generating sites, such as mitochondria41. The presence of disrupted cristae in mitochondria of Nov and Pif treated ovules (Fig. 5D) further confirms the induction of autophagic pathways. Thus inhibition of HSPs leads to failure in maintenance of homeostasis that pushes developing fiber cells to death by autophagy.
The inhibition of HSP proteins does cause a drastic change in the transcript profile of cotton fiber (Fig. 6 and Supplementary Fig. S6). Interestingly, both the inhibitors seem to target same pathways and genes, as seen by shared DEGs in both the inhibitors (Fig. 6). Thus, results indicate that HSP70 and HSP90 may have many common targets, which could be the master regulators of transcription during fiber development. Inhibition of HSPs has been reported to induce oxidative stress in the living system57. Our transcriptome data suggest that inhibitor-treated cotton ovules result in differential regulation of several known stress-related genes. AP2/EREBP transcription factors have been reported to regulate developmental, physiological and biochemical responses during different stress conditions in plants48. AP2/EREBP transcription factors, like CRF2 (cold inducible), DREB1D (dehydration and cold-inducible)48 were identified as up-regulated in our analysis whereas RAP2.3 was down-regulated59. The C3H type transcription factor such as, CZF1 which is a salt inducible transcription factor60 was identified as up-regulated in our transcriptome data. Several NAC family transcription factors that are reported in stress response61 were also up-regulated in our data. Ethylene signal transduction is crucial for fiber development as well as in stress response, ethylene signaling related genes such as ERF5 and ERF962,63 were seen up-regulated in the present study. Similarly, WRKY53 was identified as up-regulated in the transcriptome (Supplementary Fig. S7) which has been reported to be overexpressing during drought stress in Arabidopsis49. Members of C2H2 zinc finger family are also identified as up-regulated, such as ZAT10 and ZAT12 (cold inducible). We observed up-regulation of the genes belonging to ABA metabolic pathways known to have a critical role in stress hormone in plants64. Calcium acts as an essential signaling molecule in both fiber development and in coping with stress. Enhanced H2O2 levels also induce Ca2+ signaling pathways65, the rise in Ca2+ signaling pathways in the present study might be due to the high concentration of H2O2 in inhibitor-treated ovules (Fig. 6). All the genes and pathways that were up-regulated points towards the activation of multiple stress-related pathways during HSP inhibition. Inhibition of HSPs facilitates protein degradation via ubiquitin-mediated pathways66. In concordance, we identified up-regulation of several genes involved in ubiquitin-mediated protein degradation. Similarly, protein degradation related PHOR1 transcription factors were also up-regulated67. Thus, results indicate that HSP inhibition by treatment with inhibitors results in improper protein folding and that may lead the proteins towards ubiquitin-mediated protein degradation pathways in cotton fiber.
Apart from up-regulated genes, there were several genes that are reported to play a role in fiber elongation, such as Pectate lyase68, WRKY69, GDSL-lipase70, lignin biosynthesis related gene UGT72E171 were down-regulated in the present study. Further, Cytochrome p450 genes which are involved in brassinosteroid biosynthesis and having an important role in fiber elongation were down-regulated in our study72. Expansins are the cell wall loosening enzymes which are important for fiber elongation73 were found to be down-regulated in the present study. Thus major conclusion from transcriptome indicates that inhibition of HSPs leads to up-regulation of genes and pathways that are involved in managing stress and down-regulation of several genes reported to play an essential role in fiber development.
Thus, our study points towards the importance of HSP90 and HSP70 in fiber initiation and elongation. HSP90 and HSP70 inhibition lead to oxidative stress and autophagic cell death in initiating and elongating fiber cells. We observed up-regulation of genes belonging to several stress-related pathways and down-regulation of several fiber-elongation related genes concurrently to inhibition of HSP90 and HSP70. Our study thus points out the importance of chaperone and their possible engineering for better fiber yield and quality.
Material and Methods
Plant Materials
G. hirsutum genotype JKC725 was used in the present study for all the experimental purposes. Ovules from field grown plants were excised at −3 and 0 DPA for in-vitro ovule culture. The 6 DPA ovules were used for RNA extraction and full-length cloning of GhHSP90 and GhHSP70.
Microarray data retrieval and transcript level analysis of HSPs
Cotton fiber in-house microarray data (GSE36228) from our previous study34 was used to perform gene expression analysis at different stages of fiber development (0, 6, 9, 12, 19 and 25 DPA). Based on the annotation of probe sets, log2 expression values of all the HSP genes were fetched (p-value ≤ 0.05). A heat map was generated to visualize the transcript level of significantly expressed HSP genes using MeV v2.0 software (http://mev.tm4.org/#/welcome).
Gene family, phylogenetic tree and transcript level analysis
The Arabidopsis HSP90 and HSP70 protein sequences were used as a query to perform BlastP similarity search (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/) against G. hirsutum proteome data available at CottonGen database74. The sequences obtained were analyzed for the presence of characteristic domains of both the families using Conserved Domain Database (CDD; https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi)75. Further, the identified genes were classified as A- and D-subgenome homeologues by using their homology to respective subgenomes. The identified HSP genes were assigned name following the nomenclature pattern in the previous publication26. For sub-cellular localization prediction two web-based tools, namely, CELLOv.2.5 and TargetP 1.1 were used. The protein sequence of all the members was aligned using inbuilt ClustalW program of MEGA v6.06 package. HSP90, and HSP70 specific phylogenetic analysis was performed using MEGA v6.06 and the neighbor-joining method with 1000 bootstrap value76. To examine the transcript levels of the HSP90 and HSP70 gene family members in different fiber developmental stages, publically available transcriptome datasets5 were downloaded from NCBI SRA database (Supplementary Table S1) and analyzed using DNASTAR QSeq software77. The expression profile of genes was visualized using a heatmap generated using MeV v2.0 software78.
In-vitro cotton ovule culture and inhibitor treatment
In-vitro ovule culture was performed with cotton ovules excised at −3 DPA for initiation and 0 DPA for elongation related studies using the method described by Beasley and Ting (1973)79. In brief, the ovules were surface sterilized with 0.1% HgCl2 solution (w/v) and cultured in ½ murashige and skoog liquid (MSL) media supplemented with plant growth hormones, 5 µM α-NAA (Sigma Aldrich) and 0.5 µM gibberellic acid (Sigma Aldrich) and incubated at 32 °C in the dark. HSP90 and HSP70 specific inhibitors, novobiocin (Sigma Aldrich) and pifithrin (Sigma Aldrich) respectively, were used in the present study to elucidate their role in fiber development. The inhibitors were dissolved in dimethyl sulfoxide (DMSO; Sigma Aldrich) and added to cultured ovules at various concentrations (Nov: 6.3, 15.7 and 31.5 µM and Pif: 16, 20 and 24 µM). The effect of HSP inhibition on initiation, elongation and SCW stage, inhibitor treatment was observed on cultured ovules in six biological and three technical replicates at −3, 3 and 14 DPA, respectively. An equal concentration of DMSO was used to treat the control ovules. Cultured ovules were examined for the difference in fiber development under control and inhibited conditions. The inhibitor concentration (IC50) value that corresponds to 50% fiber growth inhibition was calculated for both the inhibitors by estimating their TFU. Further, images were taken at 6, 12 and 24 DPA for initiation, elongation and SCW stages, respectively, using Lumix DMC FZ-70 camera (Panasonic).
Scanning Electron Microscopy
Control and treated in-vitro cultured ovules at 1 DPA stage were washed twice with 1XPBS (pH 7.2) followed by thorough washing in 0.1 M sodium cacodylate buffer and fixed in 2.5% glutaraldehyde and 4% paraformaldehyde solution for overnight at 4 °C. Ovules were again washed thrice using 0.1 M sodium cacodylate for 20 min each and then transferred in osmium tetraoxide for overnight. Further, two washings of 0.1 M sodium cacodylate were conducted to remove excess osmium tetraoxide. Next, dehydration was carried out in acetone series using 15%, 30%, 60% and 90% solution. At least 3 changes were made in 100% acetone for 20 min each. Samples were dehydrated till they reach critical point of dehydration (CPD) and finally coated with platinum particles (2 coating). The platinum coated samples were observed under the scanning electron microscope (FEG450 Quanta, Netherland).
Quantitative estimation of fiber parameters
To observe the effect of inhibitors on fiber growth of in-vitro cultured 3 DPA old ovules (initiation stage) and 12 DPA old ovules (elongation stage) was analyzed by estimating TFU80. Briefly, treated and control ovules in three biological and three technical replicates were stained in toluidine blue solution, followed by through washing with distilled water. The stained ovules were immersed in the de-staining solution and absorbance of the de-staining solution was monitored (in triplicates) at 624 nm on UV-Vis spectrophotometer (Shimadzu, Japan) after 1 h incubation. The concentration at which the TFU value was half to that of control ovules was designated as IC50 value for the corresponding inhibitor. Further, to examine the effect of inhibitors in SCW stage, cellulose content was estimated in control and treated ovules at 24 DPA using Anthrone method81.
Bacterial expression and purification of recombinant GhHSP90-7 and GhHSP70-8 in E. coli
Bacterial expression of recombinant GhHSP90-7 and GhHSP70-8 was performed using champion pET-SUMO expression system (Invitrogen). The full-length coding sequence of GhHSP90-7 (2.1 Kb) and GhHSP70-8 (1.8 Kb) was amplified with advantage Taq DNA polymerase (Clontech) using gene-specific primers (Supplementary Table S4) and cloned in the pET-SUMO TA-cloning vector. E. coli BL21 (RIL) strain was used to express the proteins. Bacterially expressed recombinant proteins were purified using Ni-NTA columns (Qiagen) according to the manufacturer’s protocol and were confirmed on SDS-PAGE followed by immunoblot using anti penta-His antibody (Qiagen).
Molecular chaperone assay
The chaperone activity of HSP90 and HSP70 proteins was measured by incubating with substrate CS at an elevated temperature, and the first reaction of the citric acid cycle was monitored35. In the first reaction of citric acid cycle acetyl-CoA reacts with oxaloacetic acid in the presence of CS to form acetyl-CoA thioester. DTNB oxidizes acetyl-CoA thioester to form a yellow product that is observed spectrophotometrically. The aggregation of 0.5 µM citrate synthase (from Porcine heart; Sigma) was induced at 43 °C in 50 mM HEPES (pH 8.0), with or without 1.8 µM proteins (BSA or HSP90/70 (Human, Sigma) or GhHSP90/70) and with or without HSP90/70 inhibitors Nov or Pif respectively. The activity of CS was monitored spectrophotometrically at 412 nm in the presence of 0.1 mM oxalo acetic acid, 0.1 mM DTNB [5,5’-dithiobis(2-nitrobenzoic acid)] and 0.05 mM acetyl-CoA in TE buffer (50 mM tris, 2 mM EDTA, pH 8.0). The readings recorded at 25 °C at an interval of 10 min for total of 40 min. The experiment was performed twice, each with two biological and three technical replicates.
Histochemical detection of relative H2O2 and superoxide in cultured ovules
Relative H2O2 levels were measured in control and inhibitor-treated ovules at 0 and 6 DPA, using cell-permeable 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA)82. Control and inhibitor-treated cotton ovules in three biological and two technical replicates were incubated in H2DCFDA solution (0.5 mg/ml in 1XPBS) at room temperature. Conversion of non-fluorescent H2DCFDA to fluorescent 2′,7′-dichlorofluorescein (DCF) in ovules was measured by fluorimeter (BioTek FLX800; excitation/emission: 485/525 nm) for 60 min at the interval of 10 min. For the detection of superoxide radicals, control and treated ovules at 0 and 6 DPA were stained with nitro-blue tetrazolium chloride (NBT) (Roche Diagnostics)83. Ovules were stained with NBT staining solution (0.2% NBT in 50 mM sodium phosphate buffer) for 10 min. Followed by through washing with distilled water. The images of ovules were captured under stereo-microscope (Leica MZ 125, Germany).
Ascorbate peroxidase assay
The control and inhibitor-treated cultured ovules at 0 DPA were crushed in liq. nitrogen and homogenized in 1 ml extraction buffer containing 50 mM phosphate buffer (pH 7.0), 1 mM EDTA, 2% PVP, 10% glycerol and 1 mM ascorbate. The homogenate was centrifuged at 13,000xg for 30 min at 4 °C and supernatant was used for estimation of APX activity by spectrophotometric method7,84. Briefly, 100 µl of sample was mixed with assay buffer containing 50 mM phosphate buffer (pH 7.0), 0.1 mM EDTA and 0.5 mM ascorbate. The reaction was initiated by addition of 0.1 mM H2O2 and change in absorbance was monitored at 290 nm in UV/Vis-spectrophotometer (Perkin-Elmer) for 3 min at 30 seconds interval. The APX activity was calculated by a decrease in absorbance of ascorbate. One unit of enzyme activity was defined as the amount of APX required for the oxidization of 1 μmol ascorbate at 25 °C in 1 min.
MDC staining
MDC dye was used to stain autophagic vesicles. Ovules at 0 and 1 DPA were stained with a 0.05 mM final concentration of monodansylcadaverine (Sigma) in 1XPBS for 10 min85. Ovules were washed twice with 1XPBS to remove excess MDC. Transverse sections of ovule were observed under LSM 510 META confocal microscope (Carl Zeiss), with an excitation wavelength of 335 nm and an emission band pass of 505–535 nm.
Transmission electron microscopy
The ovules were cultured at −3 DPA and treated with inhibitors. At 1 DPA the control and treated ovules were washed with 1XPBS (pH 7.2). The ovules were then fixed in 2.5% glutaraldehyde prepared in 0.1 M sodium cacodylate buffer (pH 7.2) (Ladd Research) for 4 h at 4 °C followed by three times washing with 0.1 M sodium cacodylate buffer. Further, the ovules were treated with 1% osmium tetraoxide for 4 h and thoroughly washed with sodium cacodylate. After this, the ovules were dehydrated in acetone series (15–100%) and then embedded in araldite-DDSA mixture (Ladd Research Industries, USA) followed by baking at 60 °C. Ultra-microtome (Leica EM UC7) was used to cut 60-80 nm thick sections from the blocks. Further, these sections were stained with uranyl acetate and lead citrate and analyzed under FEI Tecnai G2spirit twin transmission electron microscope equipped with Gatan digital CCD camera (Netherland) at 80 kV.
RNA extraction from cultured ovules
Total RNA was isolated from control (DMSO), HSP90 and HSP70 inhibitor-treated ovules at 6 DPA using the spectrum plant total RNA isolation kit (Sigma Aldrich). After DNaseI (Ambion) treatment, quality and quantity of samples were checked using 2100 Agilent Bioanalyzer (Agilent) and Nanodrop 1000 spectrophotometer (Thermo Scientific), respectively.
Transcriptome analysis
Control and inhibitor treated RNA samples at 6 DPA were used for library preparation and sequenced by paired end sequencing method14 with Illumina NextSeq. 500 sequencing platform. The fastq files generated after sequencing were quality filtered using FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) (Supplementary Table S2). The processed reads were aligned with default parameters using Tophat (version 2.1.1) on G. hirsutum genome downloaded from Cotton Genome Project (CGP) (http://cgp.genomics.org.cn/page/species/download.jsp?category=hirsutum). The estimation of FPKM (Fragment per kilo per million) values for expression of genes and transcripts were performed using Cufflinks program (version 2.0.0) with default parameters. The DEGs were filtered by p-value ≤0.01, FDR ≤0.05 and log fold change > 1, and these DEGs were used further for pathway analysis using Mapman (version 3.5.1R2).
qRT-PCR Analysis
First-strand cDNA was synthesized from DNaseI treated total RNA using SuperScript III (Invitrogen). The qRT-PCR reaction was carried out for selected DEGs (Supplementary Table S4) using SYBR Green PCR master mix (Invitrogen) in ABI7500 Fast real-time PCR system (Applied Biosystems). All the reactions were performed in three biological and three technical replicates. GbUbiQ1 and Histone3 (Accession number AY375335 and AF024716, respectively)86 were used as internal controls for data normalization. Further, average fold change was calculated from the normalized data by using the ∆∆Ct method of ABI7500 SDS software (version 1.2.2).
Data Availability
Sequence data generated for this study will be available with the NCBI SRA database under the bioproject accession number PRJNA397595.
Electronic supplementary material
Acknowledgements
We thank Dr. Sunil K. Singh for his help in ovule culture and biochemical assays; Ms. Poonam Pant for help in transcriptome data analysis. We also acknowledge support of Dr. P.N. Saxena for SEM and Mr. Jai Shankar for TEM imaging at Electron Microscopy facility, CSIR-IITR, Lucknow. This work was supported by grants (BSC0107) from Council of Scientific and Industrial Research (CSIR), India.
Author Contributions
S.V.S. conceived the idea. A.S. and S.V.S. planned the work. A.S. executed most of the experiments. K.M.R. helped in family analysis and data interpretation, and performed phylogenetic and in-silico gene expression study. A.C. helped with biochemical assays and ovule culture. V.K.Y. performed confocal microscopy. S.K.A. helped in data interpretation. A.S. and S.V.S. analyzed the data and wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21866-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stiff, M. R. & Haigler, C. H. Recent advances in cotton fiber development. Flowering and fruiting in cotton. Tennessee: The Cotton Foundation, 163-192 (2012).
- 2.Lee JJ, Woodward AW, Chen ZJ. Gene expression changes and early events in cotton fibre development. Annals of botany. 2007;100:1391–1401. doi: 10.1093/aob/mcm232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruan Y-L, Chourey PS, Delmer DP, Perez-Grau L. The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiology. 1997;115:375–385. doi: 10.1104/pp.115.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan J, et al. A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant physiology. 2013;162:86–95. doi: 10.1104/pp.112.212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D, Zhang T, Guo W. Effect of H 2 O 2 on fiber initiation using fiber retardation initiation mutants in cotton (Gossypium hirsutum) Journal of plant physiology. 2010;167:393–399. doi: 10.1016/j.jplph.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Qin Y-M, Hu C-Y, Zhu Y-X. The ascorbate peroxidase regulated by H2O2 and ethylene is involved in cotton fiber cell elongation by modulating ROS homeostasis. Plant signaling & behavior. 2008;3:194–196. doi: 10.4161/psb.3.3.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li HB, et al. A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytologist. 2007;175:462–471. doi: 10.1111/j.1469-8137.2007.02120.x. [DOI] [PubMed] [Google Scholar]
- 8.Guo K, et al. Fibre elongation requires normal redox homeostasis modulated by cytosolic ascorbate peroxidase in cotton (Gossypium hirsutum) Journal of experimental botany. 2016;67:3289–3301. doi: 10.1093/jxb/erw146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gechev TS, Hille J. Hydrogen peroxide as a signal controlling plant programmed cell death. The Journal of cell biology. 2005;168:17–20. doi: 10.1083/jcb.200409170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houot V, et al. Hydrogen peroxide induces programmed cell death features in cultured tobacco BY‐2 cells, in a dose‐dependent manner. Journal of Experimental Botany. 2001;52:1721–1730. [PubMed] [Google Scholar]
- 11.Katschinski DM, Boos K, Schindler SG, Fandrey J. Pivotal role of reactive oxygen species as intracellular mediators of hyperthermia-induced apoptosis. Journal of Biological Chemistry. 2000;275:21094–21098. doi: 10.1074/jbc.M001629200. [DOI] [PubMed] [Google Scholar]
- 12.Lindquist S. The heat-shock response. Annual review of biochemistry. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 13.De Maio A. Heat shock proteins: facts, thoughts, and dreams. Shock. 1999;11:1–12. doi: 10.1097/00024382-199901000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Koning AJ, Rose R, Comai L. Developmental expression of tomato heat-shock cognate protein 80. Plant physiology. 1992;100:801–811. doi: 10.1104/pp.100.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marrs KA, et al. Characterization of two maize HSP90 heat shock protein genes: expression during heat shock, embryogenesis, and pollen development. genesis. 1993;14:27–41. doi: 10.1002/dvg.1020140105. [DOI] [PubMed] [Google Scholar]
- 16.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 17.Hahn A, Bublak D, Schleiff E, Scharf K-D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. The Plant Cell. 2011;23:741–755. doi: 10.1105/tpc.110.076018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X-P, Glaser E. Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends in plant science. 2002;7:14–21. doi: 10.1016/S1360-1385(01)02180-X. [DOI] [PubMed] [Google Scholar]
- 19.Hafrén A, Hofius D, Rönnholm G, Sonnewald U, Mäkinen K. HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. The Plant Cell. 2010;22:523–535. doi: 10.1105/tpc.109.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, Schöffl F. AnHsp70 antisense gene affects the expression of HSP70/HSC70, the regulation of HSF, and the acquisition of thermotolerance in transgenicArabidopsis thaliana. Molecular and General Genetics MGG. 1996;252:11–19. doi: 10.1007/s004389670002. [DOI] [PubMed] [Google Scholar]
- 21.Su P-H, Li H-m. Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant physiology. 2008;146:1231–1241. doi: 10.1104/pp.107.114496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jungkunz I, et al. AtHsp70‐15‐deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. The Plant Journal. 2011;66:983–995. doi: 10.1111/j.1365-313X.2011.04558.x. [DOI] [PubMed] [Google Scholar]
- 23.Clément M, et al. The cytosolic/nuclear HSC70 and HSP90 molecular chaperones are important for stomatal closure and modulate abscisic acid-dependent physiological responses in Arabidopsis. Plant physiology. 2011;156:1481–1492. doi: 10.1104/pp.111.174425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodziewicz P, Swarcewicz B, Chmielewska K, Wojakowska A, Stobiecki M. Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiologiae Plantarum. 2014;36:1–19. doi: 10.1007/s11738-013-1402-y. [DOI] [Google Scholar]
- 25.Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulatory network of plant heat stress response. Trends in Plant Science. 2017;22:53–65. doi: 10.1016/j.tplants.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 26.Sarkar NK, Kundnani P, Grover A. Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa) Cell Stress and Chaperones. 2013;18:427–437. doi: 10.1007/s12192-012-0395-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishna P, Gloor G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell stress & chaperones. 2001;6:238–246. doi: 10.1379/1466-1268(2001)006<0238:THFOPI>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, et al. Hsf and Hsp gene families in Populus: genome-wide identification, organization and correlated expression during development and in stress responses. BMC genomics. 2015;16:181. doi: 10.1186/s12864-015-1398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao P-M, et al. Proteomic identification of differentially expressed proteins in the Ligon lintless mutant of upland cotton (Gossypium hirsutum L.) Journal of proteome research. 2009;9:1076–1087. doi: 10.1021/pr900975t. [DOI] [PubMed] [Google Scholar]
- 30.Cassel JA, Ilyin S, McDonnell ME, Reitz AB. Novel inhibitors of heat shock protein Hsp70-mediated luciferase refolding that bind to DnaJ. Bioorganic & medicinal chemistry. 2012;20:3609–3614. doi: 10.1016/j.bmc.2012.03.067. [DOI] [PubMed] [Google Scholar]
- 31.Wang, R. et al. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nature communications7 (2016). [DOI] [PMC free article] [PubMed]
- 32.Allan RK, Mok D, Ward BK, Ratajczak T. Modulation of chaperone function and cochaperone interaction by novobiocin in the C-terminal domain of Hsp90 evidence that coumarin antibiotics disrupt Hsp90 dimerization. Journal of Biological Chemistry. 2006;281:7161–7171. doi: 10.1074/jbc.M512406200. [DOI] [PubMed] [Google Scholar]
- 33.Leu J-J, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Molecular cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nigam D, et al. Transcriptome dynamics during fibre development in contrasting genotypes of Gossypium hirsutum L. Plant Biotechnology Journal. 2014;12:204–218. doi: 10.1111/pbi.12129. [DOI] [PubMed] [Google Scholar]
- 35.Buchner J, Grallert H, Jakob U. [27] Analysis of chaperone function using citrate synthase as nonnative substrate protein. Methods in enzymology. 1998;290:323–338. doi: 10.1016/S0076-6879(98)90029-5. [DOI] [PubMed] [Google Scholar]
- 36.Lee GJ. Assaying proteins for molecular chaperone activity. Methods in cell biology. 1995;50:325–334. doi: 10.1016/S0091-679X(08)61040-7. [DOI] [PubMed] [Google Scholar]
- 37.Grellet Bournonville CF, Díaz‐Ricci JC. Quantitative determination of superoxide in plant leaves using a modified NBT staining method. Phytochemical Analysis. 2011;22:268–271. doi: 10.1002/pca.1275. [DOI] [PubMed] [Google Scholar]
- 38.Martin MV, Fiol DF, Sundaresan V, Zabaleta EJ, Pagnussat GC. Oiwa, a female gametophytic mutant impaired in a mitochondrial manganese-superoxide dismutase, reveals crucial roles for reactive oxygen species during embryo sac development and fertilization in Arabidopsis. The Plant Cell. 2013;25:1573–1591. doi: 10.1105/tpc.113.109306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theodoraki MA, Caplan AJ. Quality Control and Fate Determination of Hsp90 Client Proteins. Biochimica et Biophysica Acta. 2012;1823:683–688. doi: 10.1016/j.bbamcr.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin J, et al. Hydrogen peroxide-induced oxidative stress activates NF-[small kappa]B and Nrf2/Keap1 signals and triggers autophagy in piglets. RSC Advances. 2015;5:15479–15486. doi: 10.1039/C4RA13557A. [DOI] [Google Scholar]
- 41.Minibayeva F, Dmitrieva S, Ponomareva A, Ryabovol V. Oxidative stress-induced autophagy in plants: The role of mitochondria. Plant Physiology and Biochemistry. 2012;59:11–19. doi: 10.1016/j.plaphy.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Padmalatha KV, et al. Functional genomics of fuzzless-lintless mutant of Gossypium hirsutum L. cv. MCU5 reveal key genes and pathways involved in cotton fibre initiation and elongation. BMC genomics. 2012;13:624–624. doi: 10.1186/1471-2164-13-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Y, et al. Brassinosteroid regulates fiber development on cultured cotton ovules. Plant & cell physiology. 2005;46:1384–1391. doi: 10.1093/pcp/pci150. [DOI] [PubMed] [Google Scholar]
- 44.Qiu CX, et al. Computational identification of microRNAs and their targets in Gossypium hirsutum expressed sequence tags. Gene. 2007;395:49–61. doi: 10.1016/j.gene.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 45.Al-Ghazi Y, Bourot S, Arioli T, Dennis ES, Llewellyn DJ. Transcript profiling during fiber development identifies pathways in secondary metabolism and cell wall structure that may contribute to cotton fiber quality. Plant & cell physiology. 2009;50:1364–1381. doi: 10.1093/pcp/pcp084. [DOI] [PubMed] [Google Scholar]
- 46.Larkindale J, Knight MR. Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 2002;128:682–695. doi: 10.1104/pp.010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghanashyam C, Jain M. Role of auxin-responsive genes in biotic stress responses. Plant signaling & behavior. 2009;4:846–848. doi: 10.4161/psb.4.9.9376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 1819;86-96:2012. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 49.Sun Y, Yu D. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant cell reports. 2015;34:1295–1306. doi: 10.1007/s00299-015-1787-8. [DOI] [PubMed] [Google Scholar]
- 50.Sangster TA, et al. Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PloS one. 2007;2:e648. doi: 10.1371/journal.pone.0000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miemyk J. The 70 kDa stress-related proteins as molecular chaperones. Trends in plant science. 1997;2:180–187. doi: 10.1016/S1360-1385(97)85224-7. [DOI] [Google Scholar]
- 52.Barany I, Testillano PS, Mityko J, Risueño M. The switch of the microspore developmental program in Capsicum involves HSP70 expression and leads to the production of haploid plants. International Journal of Developmental Biology. 2001;45:S39–S40. [Google Scholar]
- 53.Ding M, et al. Gene expression profile analysis of Ligon lintless-1 (Li1) mutant reveals important genes and pathways in cotton leaf and fiber development. Gene. 2014;535:273–285. doi: 10.1016/j.gene.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, et al. Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.) Frontiers in plant science. 2015;6:773. doi: 10.3389/fpls.2015.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, et al. Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC genomics. 2013;14:532. doi: 10.1186/1471-2164-14-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annual review of biochemistry. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 57.Madrigal-Matute J, et al. HSP90 inhibition by 17-DMAG attenuates oxidative stress in experimental atherosclerosis. Cardiovascular research. 2012;95:116–123. doi: 10.1093/cvr/cvs158. [DOI] [PubMed] [Google Scholar]
- 58.Zhou G, et al. Silencing OsHI‐LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. The Plant Journal. 2009;60:638–648. doi: 10.1111/j.1365-313X.2009.03988.x. [DOI] [PubMed] [Google Scholar]
- 59.Dietz K-J, Vogel MO, Viehhauser A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma. 2010;245:3–14. doi: 10.1007/s00709-010-0142-8. [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, et al. Expression profile of early responsive genes under salt stress in upland cotton (Gossypium hirsutum L.) Plant Molecular Biology Reporter. 2011;29:626–637. doi: 10.1007/s11105-010-0269-y. [DOI] [Google Scholar]
- 61.Fang Y, You J, Xie K, Xie W, Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Molecular Genetics and Genomics. 2008;280:547–563. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- 62.Moffat CS, et al. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS One. 2012;7:e35995. doi: 10.1371/journal.pone.0035995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maruyama Y, et al. The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant science. 2013;213:79–87. doi: 10.1016/j.plantsci.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 64.Zhang J, Jia W, Yang J, Ismail AM. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Research. 2006;97:111–119. doi: 10.1016/j.fcr.2005.08.018. [DOI] [Google Scholar]
- 65.Mori IC, Schroeder JI. Reactive oxygen species activation of plant Ca2 + channels. A signaling mechanism in polar growth, hormone transduction, stress signaling, and hypothetically mechanotransduction. Plant Physiology. 2004;135:702–708. doi: 10.1104/pp.104.042069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doong H, et al. CAIR-1/BAG-3 Abrogates Heat Shock Protein-70 Chaperone Complex-mediated Protein Degradation ACCUMULATION OF POLY-UBIQUITINATED Hsp90 CLIENT PROTEINS. Journal of Biological Chemistry. 2003;278:28490–28500. doi: 10.1074/jbc.M209682200. [DOI] [PubMed] [Google Scholar]
- 67.Monte E, Amador V, Russo E, Martínez-García J, Prat S. PHOR1: A U-Box GA Signaling Component With a Role in Proteasome Degradation? Journal of Plant Growth Regulation. 2003;22:152–162. doi: 10.1007/s00344-003-0029-4. [DOI] [Google Scholar]
- 68.Wang H, et al. The essential role of GhPEL gene, encoding a pectate lyase, in cell wall loosening by depolymerization of the de-esterified pectin during fiber elongation in cotton. Plant molecular biology. 2010;72:397–406. doi: 10.1007/s11103-009-9578-7. [DOI] [PubMed] [Google Scholar]
- 69.Yang SS, et al. Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. The Plant journal: for cell and molecular biology. 2006;47:761–775. doi: 10.1111/j.1365-313X.2006.02829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yadav, V. K. et al. GhMYB1 regulates SCW stage-specific expression of the GhGDSL promoter in the fibres of Gossypium hirsutum L. Plant Biotechnology Journal, n/a-n/a, 10.1111/pbi.12706 (2017). [DOI] [PMC free article] [PubMed]
- 71.Lim E-K, Jackson RG, Bowles DJ. Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS letters. 2005;579:2802–2806. doi: 10.1016/j.febslet.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 72.Yang Z, et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New phytologist. 2014;203:437–448. doi: 10.1111/nph.12824. [DOI] [PubMed] [Google Scholar]
- 73.Ruan Y-L, Llewellyn DJ, Furbank RT. The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K + transporters and expansin. The Plant Cell. 2001;13:47–60. doi: 10.1105/tpc.13.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang T, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 75.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic acids research. 2015;43:D222–226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rai, K. M. et al. Identification, Characterization, and Expression Analysis of Cell Wall Related Genes in Sorghum bicolor (L.) Moench, aFood, Fodder, and Biofuel Crop. Frontiers in plant science 7 (2016). [DOI] [PMC free article] [PubMed]
- 78.Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27:3209–3210. doi: 10.1093/bioinformatics/btr490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beasley CA, Ting IP. The Effects of Plant Growth Substances on In vitro Fiber Development from Fertilized Cotton Ovules. American Journal of Botany. 1973;60:130–139. doi: 10.1002/j.1537-2197.1973.tb10209.x. [DOI] [Google Scholar]
- 80.Zhou Y, et al. Cotton (Gossypium hirsutum) 14‐3‐3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signalling. Plant biotechnology journal. 2015;13:269–280. doi: 10.1111/pbi.12275. [DOI] [PubMed] [Google Scholar]
- 81.Updegraff DM. Semimicro determination of cellulose inbiological materials. Analytical Biochemistry. 1969;32:420–424. doi: 10.1016/S0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 82.Potikha TS, Collins CC, Johnson DI, Delmer DP, Levine A. The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant physiology. 1999;119:849–858. doi: 10.1104/pp.119.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carol RJ, et al. A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature. 2005;438:1013. doi: 10.1038/nature04198. [DOI] [PubMed] [Google Scholar]
- 84.Nakano Y, Asada K. Purification of ascorbate peroxidase in spinach chloroplasts; its inactivation in ascorbate-depleted medium and reactivation by monodehydroascorbate radical. Plant and cell physiology. 1987;28:131–140. [Google Scholar]
- 85.Biederbick A, Kern H, Elsässer H. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. European journal of cell biology. 1995;66:3–14. [PubMed] [Google Scholar]
- 86.Tu L, et al. Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chinese Science Bulletin. 2007;52:3110–3117. doi: 10.1007/s11434-007-0461-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data generated for this study will be available with the NCBI SRA database under the bioproject accession number PRJNA397595.