Abstract
Epigenetics, a term with many meanings, is broadly defined as the study of dynamic states of the genome. Ciliates, a clade of unicellular eukaryotes, can teach us about the intersection of epigenetics and evolution due to the advantages of working with cultivable ciliate lineages plus their tendency to express extreme phenotypes such as heritable doublet morphology. Moreover, ciliates provide a powerful model for studying epigenetics given the presence of dimorphic nuclei – a somatic macronucleus and germline micronucleus – within each cell. Here, we exemplify the power of studying ciliates to learn about epigenetic phenomena. We highlight “classical” examples from morphology and physiology including cortical inheritance, mating type, and serotype. In addition, we detail molecular studies including DNA elimination; alternative processing and unscrambling; and copy number determination in model lineages. Based on the implications of such studies, we explore epigenetics as a possible functional mechanism for rapid speciation in ciliates.
Keywords: Ciliates, Cortical inheritance, DNA processing, Mating type, Seotype, Transgenerational epigenetics
Introduction
Epigenetics is a term with a broad array of definitions. The term “epigenetics” was first used by developmental biologist C.H. Waddington to describe how phenotypes arise from a genotype during development (Haig, 2007; Waddington, 1942). Today, the textbook definition of epigenetics regards changes in gene expression that do not rely on alteration of the nucleotide sequence (Burggren and Crews, 2014). Alterations of chromatin, including DNA methylation and histone modification, demonstrate the etymology of the word ‘epigenetics,’ which describes acting ‘above the genome’ (Haig, 2007). We prefer a broader definition similar to that put forth by Denise Barlow, one of the scientists involved in the discovery of genomic imprinting, who argued, “epigenetics has always been all the weird and wonderful things that can’t be explained by genetics” (McVittie, 2006). Under this umbrella, epigenetics includes phenomena whose molecular details are not yet fully understood. Thus is the case for many of the “classical” examples of epigenetics described below. As such, this broader definition of epigenetics also includes transgenerational inheritance, epigenetic phenomena that are heritable across generations (e.g. following conjugation in ciliates), which are the focus of this manuscript.
Ciliates as models for epigenetic studies
Due to their elaborate ciliature and beauty, ciliates have been subject to study since the invention of the microscope, generating the rich data synthesized in this manuscript. Ciliates, a monophyletic clade of unicellular eukaryotes, are characterized by the presence of cilia in at least one stage of their life cycle and the presence of dimorphic nuclei, with both a somatic macronucleus and germline micronucleus within every cell (e.g. Chalker et al., 2013; Katz, 2001; Yao et al., 2002). This combination of features contributed to the discovery of many epigenetic phenomena in ciliates.
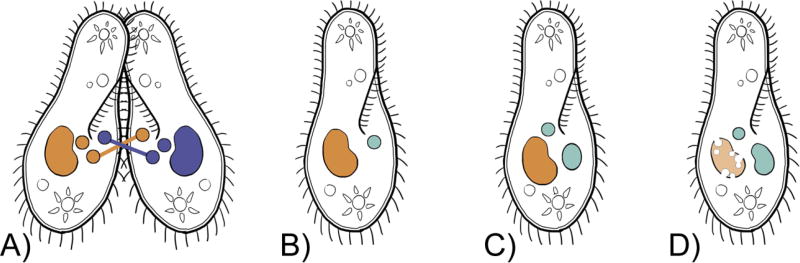
Appreciating transgenerational epigenetics’s strong impact on ciliates and their genomes requires an understanding of ciliate reproduction. During asexual reproduction, division occurs in the transcriptionally-silent micronucleus by mitosis and in the transcriptionally-active somatic macronucleus by amitosis. Sexual reproduction begins with conjugation (i.e. the fusion of two cells of different mating types) and meiosis of the micronucleus in each member of the conjugating pair (Figure 1A). Eventually, one haploid micronucleus from each cell is transferred to the other cell; the haploid nuclei then fuse within each cell to form a zygotic nucleus, which then undergoes mitosis (Chalker et al., 2013; McGrath et al., 2006; Orias, 1998). One of the two mitotic products develops into the micronucleus, presumably by heterochromatin formation that leads to a quiescent nucleus, and the other differentiates into the new macronucleus (Chalker et al., 2013; Katz, 2001; Yao et al., 2002). The DNA content and organization of the developing macronucleus are the same as the micronucleus at the beginning of macronuclear differentiation (Chalker et al., 2013). The developing macronucleus and the parental macronucleus share a cytoplasm, which allows for extensive epigenetic communication between nuclei (Chalker et al., 2013).
Fig. 1.
Simplified version of nuclear morphology following conjugation/sexual reproduction in ciliates, with the small round circles representing micronuclei: A) one haploid micronucleus is exchanged between conjugating cells; B) a zygotic nucleus is formed in the presence of the parental macronucleus; C–D) the zygotic nucleus undergoes division and daughter nuclei differentiate into a new micronucleus and macronucleus; D) the parental macronucleus degrades and is replaced by the new macronucleus.
In ciliates, inheritance following sexual reproduction is guided by a genome scanning process instead of pure Mendelian inheritance. If Mendelian inheritance were the rule in ciliate sexual reproduction, the parental macronucleus would have no effect on the developing macronucleus. However, this is not the case. While the old macronucleus degrades and contributes no DNA to the new macronucleus, it does influence the development of the new macronucleus through epigenetic processes like RNA-guided genome scanning (Baranasic et al., 2014; Chalker et al., 2013; Chalker and Yao, 2011; Meyer and Duharcourt, 1996; Nowacki and Landweber, 2009). It is possible, however, that only a portion of the genome is scanned (Hamilton et al., 2016; Schoeberl et al., 2012).
Though the molecular mechanisms underlying epigenetic processes vary, RNA-mediated processes are emerging as a common theme across eukaryotic lineages, including ciliate lineages (Fedoroff, 2012; Maurer-Alcala and Katz, 2015; Yao et al., 2002). RNA-mediated processes (referred to here as genome scanning) involve epigenetic changes to genomes regulated by single-stranded RNAs that silence transposable elements (Chalker et al., 2013; Coyne et al., 2012; Fedoroff, 2012), generate heterochromatin (Chalker and Yao, 2011), and eliminate DNA (Fang et al., 2012; Maurer-Alcala and Katz, 2015; Swart et al., 2014).
Here we highlight the roles that ciliates have played in our understanding of epigenetics over the past 50 years. We organize the manuscript around two major categories of epigenetics in ciliates: 1) “classical” examples of morphological and physiological phenomena, and 2) molecular processes (Table 1). We place ciliate epigenetic phenomena in the context of other eukaryotes to suggest that ciliates have elaborated on an ancient toolbox evolved for silencing transposons. We end by arguing that epigenetic processes in ciliates allow for increased phenotypic variation and genetic diversity.
Table 1.
Summary of examples of epigenetics in ciliates
| Classical Examples | Pattern | Processes | References |
| Cortical inheritance | Doublet morphology | Unknown | Beisson and Sonneborn (1965); Christopher and Sundermann (1995); Grimes (1973); Landman (1993) |
| Mating types | Cytoplasmic (O & E) & karyondial inheritance | Genome rearrangement | Cervantes et al. (2013); Hall and Katz (2011); Phadke and Zufall (2009); Sonneborn (1977) |
| Serotypes | Surface antigen A | Genome scanning | Baranasic et al. (2014); Beale (1957); Simon and Schmidt (2007); Sonneborn (1943) |
| Molecular Examples | |||
| DNA elimination | Removal of micronuclear-limited sequences during development | Genome scanning | Duharcourt et al. (1995); Fang et al. (2012); Hamilton et al. (2016); Jonsson et al. (2009); Swart and Nowacki (2015) |
| Alternative processing | Only found in ciliates with extensively processed macronuclear genomes | Genome scanning | Gao et al. (2015); Katz and Kovner (2010); Nowacki et al. (2011); Riley and Katz (2001) |
| Copy number determination | Inheritance of somatic copy number of nanochromosomes | Genome scanning | Bellec and Katz (2012); Heyse et al. (2010); Huang and Katz (2014); Xu et al. (2012) |
Part I: Classical examples from morphology and physiology
Classical examples of epigenetic phenomena in ciliates include cortical inheritance, and determination of mating types and serotypes. We define these phenotypic changes as examples based on morphology and physiology in that their discovery relied on light microscopy and behavioral observations.
Cortical inheritance
The inheritance of acquired changes in morphology, also known as cortical inheritance, is an epigenetic phenomenon first observed in ciliates over fifty years ago (Beisson, 2008; Beisson and Sonneborn, 1965). One of the most well studied example of cortical inheritance is doublet morphology, which refers to the acquisition and maintenance of a second, mirrored set of ciliature such as the presence of two mouths (Figure 2A, B; Bell et al., 2008; Christopher and Sundermann, 1995; Frankel and Nelsen, 1986; Landman, 1993). Doublets are produced for a variety reasons and are an example of a phenotypic change that may not require changes in the nucleotide sequences. Though janus mutant doublets may be induced genetically in Tetrayhemena thermophila (Christopher and Sundermann, 1995; Frankel and Nelsen, 1986). Doublet morphology follows Lamarck’s ideas on the inheritance of acquired characteristics. That is, when morphological changes occur, either through errors in cell division or by experimental perturbation, they are transmitted to progeny across asexual and sexual cycles (Landman, 1993). Despite the compelling data on cortical inheritance, Larmarck’s views on inheritance have been largely rejected.
Fig. 2.
Two “classical” examples of epigenetic phenomena in ciliates. Cortical inheritance: following asexual reproduction, the wild type singlet produces progeny that are also singlets (A) while mutant doublets produce doublet offspring (B). Non-Mendelian examples of mating type determination: in karyonidal inheritance, mating type varies either stochastically or is determined by changing environmental factors (e.g. temperature, time of day; C); in cytoplasmic inheritance mating types (O and E) are maternally inherited following conjugation (D; adapted from Chalker et al., 2013; Meyer and Garnier, 2002).
An example of a naturally occurring doublet phenotype is found in the ciliate Oxytricha fallax (Class Spirotrichea). If a wild-type Oxytricha fallax fails to divide, a ‘doublet’ can form such that the ciliate has two mouths and two mirrored sets of cirri (i.e. bundles of cilia) that are maintained through cell divisions (Landman, 1993). Similarly, Pleurotricha lanceolata (Spirotrichea) can form mirror-image symmetrical doublets when subjected to a combination of heat shock and surgery (Landman, 1993). Following encystment, the doublet pattern emerged unchanged and went on to reproduce doublets clonally (Grimes, 1973). Understanding the functional implications of inheritance of doublet morphology awaits additional studies, but the inheritance of acquired changes to body plans challenges the textbook views on morphological evolution.
Mating types
Non-Mendelian inheritance of mating types is another classic example of epigenetics in ciliates. Genetically determined mating types are the basis for reproductive compatibility in many eukaryotes. For example, the molecular basis for mating types has been determined in lineages such as fungi (e.g.Haber, 2012; Klar et al., 1998) and mammals (e.g. Emmons and Lipton, 2003). In ciliates, mating types can be determined in at least three ways – synclonal, karyonidal and cytoplasmic – where the last two are epigenetically regulated (Hall and Katz, 2011; Phadke and Zufall, 2009). While synclonal inheritance follows Mendelian genetics, karyonidal and cytoplasmic inheritance are epigenetically regulated (Hall and Katz, 2011; Phadke and Zufall, 2009). Karyonidal inheritance occurs when the mating type is determined either stochastically during macronuclear development or predictably by environmental stimuli such as temperature and light exposure (Figure 2C; Cervantes et al., 2013; Hall and Katz, 2011). Cytoplasmic inheritance occurs when the mating type of the offspring reflects the parent’s phenotype, in a manner analogous to maternal inheritance in animals (Figure 2D; Hall and Katz, 2011; Phadke and Zufall, 2009; Sonneborn, 1977).
An example of cytoplasmic inheritance found in Paramecium tetraurelia (Class Oligohymenophorea) involves mating types described as odd (O) and even (E) (Chalker et al., 2013; Nowacki et al., 2011; Sonneborn, 1977). The mating type is determined by cytoplasmic inheritance during the development of the new macronucleus. The O parent will produce O offspring and the E parent will produce E offspring in a manner that is independent of the alleles carried by each cell (Figure 2D; Chalker et al., 2013; Nowacki et al., 2011; Sonneborn, 1977).
Literature repeatedly demonstrates that Mendelian rules are insufficient to explain the inheritance and evolution of mating types in ciliates. Phadke and Zufall (2009) discuss the rapid diversification of mating types within ciliate species, with a particular emphasis on Tetrahymena, Paramecium and Euplotes species. They suggest that the mode of inheritance (e.g. synclonal, cytoplasmic or karyonidal), the number of mating types (2–12 in different species), and molecular determinants, such as pheromones that are secreted or cell-bound, are all rapidly evolving across the ciliate phylogeny (Phadke and Zufall, 2009). The factors (e.g. genetic, epigenetic, and selective) that drive the elevated rates of evolution, however, remain unclear (Phadke and Zufall, 2009).
Serotypes
Serotypes, determined by the kinds of surface proteins expressed and transported to the cell surface, are also epigenetically regulated in ciliates (Baranasic et al., 2014; Lepere et al., 2008; Simon and Schmidt, 2007). The phenotypic variation created by the various surface antigens enables recognition between cells, may protect ciliates from predators, or could be a defense mechanism against biotic and abiotic factors (Preer, 1986; Simon and Kusch, 2013; Sonneborn, 1970). Over the last 70 years, studies have shown that the inheritance of surface antigen genes follows Mendelian rules; however, the expression of the antigens is epigenetically regulated (Baranasic et al., 2014; Beale, 1957; Lepere et al., 2008; Sonneborn, 1943).
Microscopy studies revealed that surface antigen expression is not inherited in a Mendelian fashion in Paramecium (Beale, 1957). Antibodies were developed against the surface antigens and revealed that the expression of these genes was not inherited by Mendelian genetics but rather by cytoplasmic states (Beale, 1957). Beale (1957) showed that in Paramecium the production of surface antigens is determined by the environment, and previous history of the cytoplasmic state. The main conclusion from this study is that cells can produce a vast array of diverse antigens from a set of genes without changing the genotype (Beale, 1957). Numerous hypotheses have been put forth to explain Paramecium’s antigen formation (Beale, 1957); however, it was not until recent molecular studies that the details of serotype expression have been demonstrated to involve epigenetic inheritance (see below).
Part II: Molecular studies
While the epigenetic inheritance of morphological and physiological features has been observed in ciliates for decades, our understanding of mechanistic details has emerged more recently through molecular studies. We exemplify this progress through discussions of three epigenetic processes: DNA elimination, genome unscrambling/alternative processing, and copy number determination in extensively fragmenting ciliates. We describe how genome scanning is applicable to all these processes due to their use of small RNAs to direct genomic changes.
Germline soma distinction/DNA elimination
Elimination of particular germline sequences in macronuclear development is the most well understood epigenetic phenomenon in ciliates. DNA elimination in ciliates involves the removal of specific sequences from the developing macronucleus, and retention of sequences present in the old macronucleus (Jonsson et al., 2009; Schoeberl et al., 2012; Swart and Nowacki, 2015). The removed sequences, which can be repetitive elements or unique copy sequences referred to as internally eliminated sequences (IESs), are scattered throughout the micronuclear genome and may interrupt coding regions (Swart and Nowacki, 2015). Mechanisms of elimination have been studied in the classes Oligohymenophorea (Paramecium and Tetrahymena) and Spirotrichea (Oxytricha and Stylonychia). Though different in detail, both clades have provided evidence for a scanning mechanism in which elimination or retention of sequences in the developing macronucleus is directed by identity to small RNAs (Fang et al., 2012; Jonsson et al., 2009; Schoeberl et al., 2012; Swart et al., 2014).
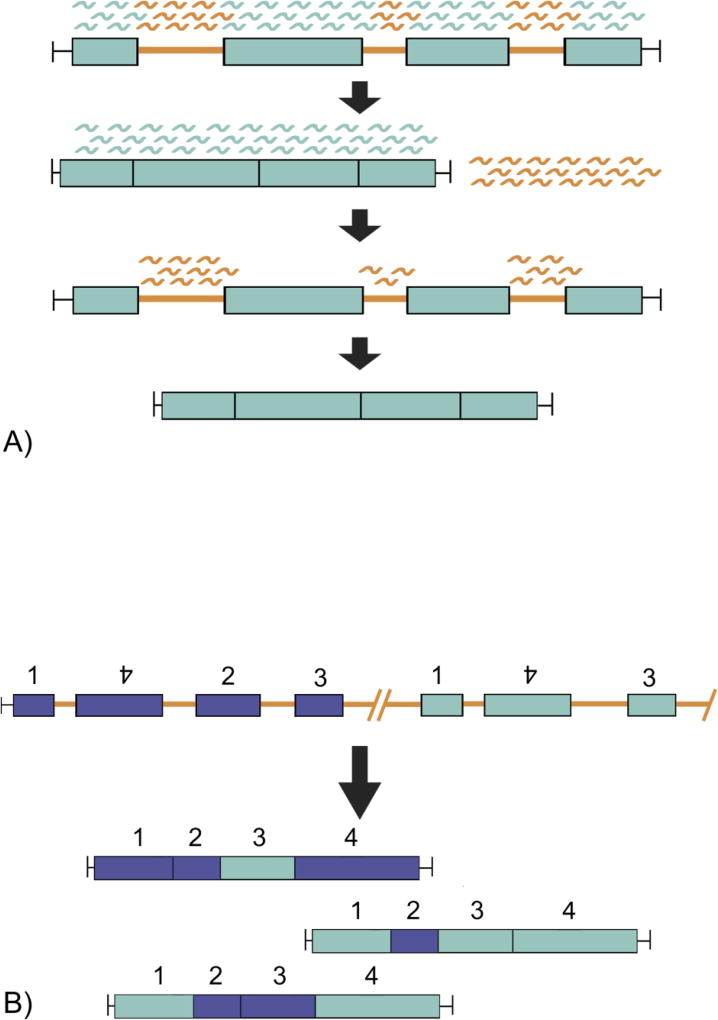
In a model proposed for Oligohymenophorea, the process of DNA elimination starts after division of the zygotic nucleus when the entire genome of the new micronucleus is transcribed (Figure 3A; Chalker and Yao, 2011; Swart et al., 2014). Recently, in Tetrahymena thermophila, micronuclear limited sequences show evidence of disproportionate representation among transcripts (Schoeberl et al., 2012). At this point, both the developing macronucleus and new micronucleus have the same genome and DNA content, so these transcripts of the micronucleus correspond to sequences in the developing macronucleus. The double-stranded RNAs (dsRNAs) are processed into small RNAs called scan RNAs (scnRNAs; Heyse et al., 2010; Swart et al., 2014). Then the scnRNAs are transported by a Piwi family protein into the old macronucleus, where they target homologous sequences (Malone and Hannon, 2009; Swart et al., 2014). The scnRNAs with homology to parental macronuclear sequences are degraded and the remaining scnRNAs are sent back into the developing macronucleus (Jonsson et al., 2009; Malone and Hannon, 2009; Swart et al., 2014). Here, the scnRNAs bind to homologous sequences not found in the parental macronucleus and these sequences are targeted for elimination in the developing macronucleus (Jonsson et al., 2009; Malone and Hannon, 2009; Swart et al., 2014). Evidence for this model was first observed in Paramecium by injecting the parental macronucleus with plasmids containing IESs (Duharcourt et al., 1995; Yao et al., 2002). About one third of these injected IESs were retained in the new macronucleus and could be passed down to future generations (Duharcourt et al., 1995; Swart et al., 2014). These studies demonstrate the importance influence of the parental macronucleus in recognizing macronuclear DNA to be retained in the next generation.
Fig. 3.
Two examples of molecular studies of epigenetic phenomena in ciliates. A) DNA elimination: in Oligohymenophorea, the entire micronuclear genome is transcribed into small scnRNAs. These transcripts are transported to the old macronucleus where they bind to regions of homology (MDSs indicated in light blue) and are degraded. The scnRNAs that did not bind to any region of the old macronucleus are transported into the new macronucleus and mark homologous regions (orange) for deletion. B) Alternative processing: MDSs from divergent paralogs are combined in varying orders to create gene variants. In the micronucleus (top) the MDSs labeled 1–4 are in a scrambled order. MDSs are reassembled and may be used in multiple macronuclear genes in alternative processing, which allows for the creation of several genes from two paralogs.
Small RNA genome scanning in Spirotrichea differs from the mechanism in Oligohymenophorea in that scnRNAs expressed in early development in Spirotrichea (referred to here as piwi-interacting RNAs or piRNAs) mark sequences for retention instead of for deletion (Chen et al., 2014; Fang et al., 2012). In Oxytricha, IESs were stably retained when injected with piRNAs homologous to these regions (Fang et al., 2012). Sequencing of piRNAs has revealed an overwhelming majority map to macronuclear destined sequences (MDS), with just 0.11% corresponding to micronuclear limited sequences (Chen et al., 2014). Fang et al. (2012) also reported that, unlike scnRNAs in Oligohymenophorea, where many scnRNAs derive from both the micronucleus and the parental macronucleus; piRNAs in Oxytricha derive only from the old macronucleus.
Differences in the function of piRNAs and scnRNAs in Spirotrichea and Oligohymenophorea respectively may be related to the extent of elimination in spirotrichs. In the model genera of Spirotrichea, Euplotes, Stylonychia and Oxytricha, around 85–98% of the sequences present in the germline are eliminated from the developing macronucleus (Fang et al., 2012; Nowacki et al., 2011; Swart and Nowacki, 2015). This value dwarfs the ~30% eliminated in Paramecium and in Tetrahymena (Arnaiz et al., 2012; Hamilton et al., 2016; Swart and Nowacki, 2015). Since the majority of germline sequences are eliminated in the Spirotrichea and the minority in Oligohymenophorea, the method of elimination for each class involves transfer of the fewest possible scnRNAs into the developing macronucleus (Fang et al., 2012).
Alternative processing and unscrambling
In a few ciliate classes, the pieces of each macronuclear gene (i.e. macronuclear destined sequences or MDSs) are not only interrupted by IESs in the micronucleus, but may also be misordered and some of the pieces may even be inverted (Chen et al., 2014; Jonsson et al., 2009; Nowacki et al., 2011). Genes assembled from misordered MDSs are referred to as scrambled, and the process of their reordering is called unscrambling (Jonsson et al., 2009; Katz and Kovner, 2010). If the same MDSs are used in the assembly of multiple macronuclear loci, the genes are described as alternatively processed (Chen et al., 2014; Jonsson et al., 2009; Nowacki et al., 2011). Gene scrambling has been reported only in ciliates with gene-sized macronuclear chromosomes, a feature present in three ciliate classes, Armophorea, Phyllopharygnea, and Spirotrichea, with possibly at least two independent origins for this process (Katz, 2001; Katz and Kovner, 2010). These ciliates share features that may support unscrambling, such as the presence of giant polytene chromosomes during macronuclear development (Ammermann, 1987; Chen et al., 2014; Katz and Kovner, 2010; Raikov, 1982).
Molecular mechanisms for unscrambling have been studied in Stylonychia and Oxytricha (Spirotrichea), where approximately 30% of macronuclear genes are scrambled in the micronucleus genomes (Jonsson et al., 2009). Long-template RNAs derived from the parental macronucleus appear to guide unscrambling (Jonsson et al., 2009; Nowacki et al., 2011; Swart and Nowacki, 2015). These long-template RNAs are theorized to serve as a scaffold for the reorganization of sequences in the developing macronucleus (Nowacki et al., 2011). The new macronuclei of cells injected with artificial RNA templates with altered orders of MDSs inherit the artificial template sequence (Jonsson et al., 2009; Nowacki et al., 2011; Nowacki et al., 2008). Long-template RNAs may also serve as a proofreading mechanism for piRNA elimination (Jonsson et al., 2009). Unscrambling occurs after initial excision, and studies of intermediates (i.e. sequences in the partially developed macronuclear genome) reveal that prior to unscrambling most elimination is imprecise, which would render the new macronucleus dysfunctional (Jonsson et al., 2009; Nowacki et al., 2011). By the time unscrambling occurs, intermediates demonstrate precise elimination, indicating a proofreading mechanism must be present (Jonsson et al., 2009).
Alternative processing is a variation on gene unscrambling that may generate protein diversity (Figure 3B; Katz and Kovner, 2010). Similar to V(D)J recombination in the human immune system, alternative processing involves the reordering of sequences in a multiplicity of arrangements to create diverse gene family members. For example, the ciliate Chilodonella uncinata (Phyllopharyngea) generates multiple macronuclear β-tubulin paralogs from an alternatively processed shared micronuclear region (Gao et al., 2015; Katz and Kovner, 2010). In Oxytricha, over 1000 MDSs are reused to generate alternatively processed genes, and one MDS can contribute to at most five gene family members (Chen et al., 2014). The ability to recycle MDSs may lead to greater diversity within protein families, as ciliates with extensively processed genomes have higher rates of protein evolution compared to non-scrambling ciliates (Gao et al., 2015; Zufall et al., 2006).
Copy number determination
Copy number of chromosomes in the macronucleus is another epigenetically controlled phenomenon in ciliates with extensively processed, gene-sized macronuclear chromosomes (Bellec and Katz, 2012; Xu et al., 2012). Ciliates with multigene macronuclear chromosomes, such as Paramecium and Tetrahymena, have roughly even copy numbers of all chromosomes in their macronuclei (Xu et al., 2012). For example, each chromosome in Paramecium tetraurelia is amplified to ~800 copies (Duret et al., 2008). In contrast, copy numbers in Oxytricha and Chilodonella range from a couple hundred up to a million for each of 15,000–20,000 different nanochromosomes (Huang and Katz, 2014; Jonsson et al., 2009).
Like unscrambling, copy number determination is thought to rely on long template RNAs from the old macronucleus (Heyse et al., 2010; Nowacki et al., 2011; Xu et al., 2012). In Stylonychia, the developing macronucleus begins with roughly the same copy number for each gene, like the zygotic nucleus from which it came (Heyse et al., 2010). Experiments using RNA interference (RNAi) against highly amplified genes demonstrated a decrease in copy number when the corresponding template was degraded (Heyse et al., 2010). Conversely, injection of template RNAs results in an increase in DNA copy number (Heyse et al., 2010; Nowacki et al., 2010). These alterations in Stylonychia were stably inherited through vegetative growth; similar results have been obtained for Oxytricha (Heyse et al., 2010; Nowacki et al., 2011).
Part III: Implications - The transposon link
A model proposed for the origin of epigenetic mechanisms in ciliates is that they are derived from transposon silencing machinery (Klobutcher and Herrick, 1997). The mechanisms underlying genome scanning in ciliates and epigenetic processes in other eukaryotic lineages share similarities with transposon excision machinery (Arnaiz et al., 2012; Chalker et al., 2013; Coyne et al., 2012; Maurer-Alcala and Katz, 2015; Schoeberl and Mochizuki, 2011; Singh et al., 2014; Swart et al., 2014). For instance, in Euplotes and Paramecium species, the TA dinucleotides flanking IESs bear resemblance to the termini of Tc1/mariner transposons (Arnaiz et al., 2012; Chalker et al., 2013; Coyne et al., 2012). Tetrahymena species use epigenetically regulated heterochromatin formation to remove IESs, similar to other eukaryotes in which small RNA-directed heterochromatin formation is used to control transposons (Chalker and Yao, 2011; Nowacki et al., 2011). Furthermore, in many ciliate lineages, key enzymes involved in the epigenetic removal of IESs are derived from transposases (Chalker et al., 2013; Coyne et al., 2012; Klobutcher and Herrick, 1997; Swart et al., 2014).
Genome scanning has been linked to at least some of the classic examples of epigenetic phenomena in ciliates; for example, both Paramecium tetraurelia and Paramecium septaurelia use genome scanning in mating type determination (one of our classical examples; Singh et al., 2014). Additionally, Tetrahymena thermophila uses programmed DNA rearrangements in mating type determination through karyonidal inheritance (Cervantes et al., 2013). Both scnRNAs and small interfering RNAs (siRNAs) have been implicated as possible epigenetic mechanisms for the inheritance of surface antigens (Baranasic et al., 2014; Lepere et al., 2008). Hence, the epigenetic processes described here likely reflect modification of genome defense machinery that originally evolved to silence invading transposable elements.
Implications - Macroevolution
Epigenetic phenomena may influence macroevolution in ciliates by contributing to rates of speciation. For examples, changes in gene copy number or locations of micronuclear-limited sequences in isolated populations may create barriers to genetic exchange, leading to rapid speciation events (Gao et al., 2015; Hall and Katz, 2011). Similarly, differential loss of duplicated macronuclear destined sequences, which can be corrected through gene unscrambling and alternative processing, may inhibit production of viable offspring between members of previously isolated populations (Gao et al., 2015). This rapid speciation as the result of epigenetic processes may contribute to the complex patterns of morphological and molecular evolution such as cryptic species, which are common in ciliates (Foissner et al., 2008; Hall and Katz, 2011; Lahr et al., 2014; Przybos and Tarcz, 2016; Sonneborn, 1975).
Acknowledgments
We thank Chip Sisson for illustrating the figures, Ying Yan for formatting the references, other members of the Katz Lab for commenting on earlier versions of this manuscript, and our reviewers for their insights. This work is supported by grants from the National Science Foundation (DEB-1541511) and the National Institutes of Health (Area 1R15GM113177-01) to L.A.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ammermann D. Giant chromosomes in ciliates. Results Probs Cell Differ. 1987;14:59–67. doi: 10.1007/978-3-540-47783-9_4. [DOI] [PubMed] [Google Scholar]
- Arnaiz O, Mathy N, Baudry C, Malinsky S, Aury JM, Wilkes CD, Garnier O, Labadie K, Lauderdale BE, Le Mouel A, Marmignon A, Nowacki M, Poulain J, Prajer M, Wincker P, Meyer E, Duharcourt S, Duret L, Betermier M, Sperling L. The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. Plos Genetics. 2012;8:e1002984. doi: 10.1371/journal.pgen.1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranasic D, Oppermann T, Cheaib M, Cullum J, Schmidt H, Simon M. Genomic Characterization of Variable Surface Antigens Reveals a Telomere Position Effect as a Prerequisite for RNA Interference-Mediated Silencing in Paramecium tetraurelia. Mbio. 2014;5:e01328–01314. doi: 10.1128/mBio.01328-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale GH. The Antigen System of Paramecium-Aurelia. Int Rev Cytol. 1957;6:1–23. [Google Scholar]
- Beisson J. Preformed cell structure and cell heredity. Prion. 2008;2:1–8. doi: 10.4161/pri.2.1.5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson J, Sonneborn TM. Cytoplasmic Inheritance of Organization of Cell Cortex in Paramecium Aurelia. P Natl Acad Sci USA. 1965;53:275–282. doi: 10.1073/pnas.53.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AJ, Satir P, Grimes GW. Mirror-imaged doublets of Tetmemena pustulata: Implications for the development of left-right asymmetry. Developmental Biology. 2008;314:150–160. doi: 10.1016/j.ydbio.2007.11.020. [DOI] [PubMed] [Google Scholar]
- Bellec L, Katz LA. Analyses of chromosome copy number and expression level of four genes in the ciliate Chilodonella uncinata reveal a complex pattern that suggests epigenetic regulation. Gene. 2012;504:303–308. doi: 10.1016/j.gene.2012.04.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggren WW, Crews D. Epigenetics in comparative biology: why we should pay attention. Integrative and Comparative Biology. 2014;54:7–20. doi: 10.1093/icb/icu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Hamilton EP, Xiong J, Lawson MJ, Yuan D, Hadjithomas M, Miao W, Orias E. Selecting one of several mating types through gene segment joining and deletion in Tetrahymena thermophila. PLoS Biol. 2013;11:e1001518. doi: 10.1371/journal.pbio.1001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Meyer E, Mochizuki K. Epigenetics of Ciliates. Cold Spring Harbor Perspectives in Biology. 2013;5 doi: 10.1101/cshperspect.a017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. DNA Elimination in Ciliates: Transposon Domestication and Genome Surveillance. In: Bassler BL, Lichten M, Schupbach G, editors. Annual Review Genetics. Vol. 45. 2011. pp. 227–246. [DOI] [PubMed] [Google Scholar]
- Chen X, Bracht JR, Goldman AD, Dolzhenko E, Clay DM, Swart EC, Perlman DH, Doak TG, Stuart A, Amemiya CT, Sebra RP, Landweber LF. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell. 2014;158:1187–1198. doi: 10.1016/j.cell.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher GK, Sundermann CA. Doublet cells in Tetrahymena as indicators of culture media composition. Biol Trace Elem Res. 1995;50:181–191. doi: 10.1007/BF02785409. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Lhuillier-Akakpo M, Duharcourt S. RNA-guided DNA rearrangements in ciliates: Is the best genome defence a good offence? Biology of the Cell. 2012;104:309–325. doi: 10.1111/boc.201100057. [DOI] [PubMed] [Google Scholar]
- Duharcourt S, Bulter A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence in Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- Duret L, Cohen J, Jubin C, Dessen P, Gout JF, Mousset S, Aury JM, Jaillon O, Noel B, Arnaiz O, Betermier M, Wincker P, Meyer E, Sperling L. Analysis of sequence variability in the macronuclear DNA of Paramecium tetraurelia: A somatic view of the germline. Genome Research. 2008;18:585–596. doi: 10.1101/gr.074534.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons SW, Lipton J. Genetic basis of male sexual behavior. J Neurobiol. 2003;54:93–110. doi: 10.1002/neu.10163. [DOI] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff NV. Presidential address. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- Foissner W, Chao A, Katz LA. Diversity and geographic distribution of ciliates (Protista: Ciliophora) Biodiversity and Conservation. 2008;17:345–363. [Google Scholar]
- Frankel J, Nelsen EM. Intracellular pattern reversal in Tetrahymena thermophila. II. Transient expression of a janus phenocopy in balanced doublets. Dev Biol. 1986;114:72–86. doi: 10.1016/0012-1606(86)90384-2. [DOI] [PubMed] [Google Scholar]
- Gao F, Roy SW, Katz LA. Analyses of alternatively processed genes in ciliates provide insights into the origins of scrambled genomes and may provide a mechanism for speciation. mBio. 2015;6:e01998–01914. doi: 10.1128/mBio.01998-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes GW. An analysis of the determinative difference between singlets and doublets of Oxytricha fallax. Genet Res. 1973;21:57–66. [Google Scholar]
- Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Weismann rules! OK? Epigenetics and the Lamarckian temptation. Biology & Philosophy. 2007;22:415–428. [Google Scholar]
- Hall MS, Katz LA. On the nature of species: insights from Paramecium and other ciliates. Genetica. 2011;139:677–684. doi: 10.1007/s10709-011-9571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EP, Kapusta A, Huvos PE, Bidwell SL, Zafar N, Tang H, Hadjithomas M, Krishnakumar V, Badger JH, Caler EV, Russ C, Zeng Q, Fan L, Levin JZ, Shea T, Young SK, Hegarty R, Daza R, Gujja S, Wortman JR, Birren BW, Nusbaum C, Thomas J, Carey CM, Pritham EJ, Feschotte C, Noto T, Mochizuki K, Papazyan R, Taverna SD, Dear PH, Cassidy-Hanley DM, Xiong J, Miao W, Orias E, Coyne RS. Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. Elife. 2016;5 doi: 10.7554/eLife.19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyse G, Jonsson F, Chang WJ, Lipps HJ. RNA-dependent control of gene amplification. P Natl Acad Sci USA. 2010;107:22134–22139. doi: 10.1073/pnas.1009284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Katz LA. Nanochromosome Copy Number Does not Correlate with RNA Levels Though Patterns are Conserved between Strains of the Ciliate Morphospecies Chilodonella uncinata. Protist. 2014;165:445–451. doi: 10.1016/j.protis.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson F, Postberg J, Lipps HJ. The unusual way to make a genetically active nucleus. DNA and Cell Biology. 2009;28:71–78. doi: 10.1089/dna.2008.0806. [DOI] [PubMed] [Google Scholar]
- Katz LA. Evolution of nuclear dualism in ciliates: a reanalysis in light of recent molecular data. Int. J. Syst. Evol. Microbiol. 2001;51:1587–1592. doi: 10.1099/00207713-51-4-1587. [DOI] [PubMed] [Google Scholar]
- Katz LA, Kovner AM. Alternative processing of scrambled genes generates protein diversity in the ciliate Chilodonella uncinata. J. Exper. Zool. B. 2010;314:480–488. doi: 10.1002/jez.b.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJS, Ivanova AV, Dalgaard JZ, Bonaduce MJ, Grewal SIS. Multiple epigenetic events regulate mating-type switching of fission yeast. Epigenetics. 1998;214:87–99. doi: 10.1002/9780470515501.ch6. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Herrick G. Developmental genome reorganization in ciliated protozoa: The transposon link. Progress in Nucleic Acid Research and Molecular Biology. 1997;56:1–62. doi: 10.1016/s0079-6603(08)61001-6. [DOI] [PubMed] [Google Scholar]
- Lahr DJG, Laughinghouse HDI, Gao F, Oliverio A, Katz LA. How discordant morphological and molecular evolution among microorganisms can revise our notions of biodiversity on Earth. Bioessays. 2014;36:950–959. doi: 10.1002/bies.201400056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman OE. Inheritance of acquired characteristics revisited. Scientific American. 1993;43:696–705. [Google Scholar]
- Lepere G, Betermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as Guardians of the Genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Alcala XX, Katz LA. An epigenetic toolkit allows for diverse genome architectures in eukaryotes. Current Opinion in Genetics & Development. 2015;35:93–99. doi: 10.1016/j.gde.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath C, Zufall RA, Katz LA. Genome evolution in ciliates. In: Katz LA, Bhattacharya D, editors. Genomics and Evolution of Eukaryotic Microbes. Oxford University Press; 2006. [Google Scholar]
- McVittie B. What is Epigenetics. © Epigenome NoE 2006 [Google Scholar]
- Meyer E, Duharcourt S. Epigenetic programming of developmental genome rearrangements in ciliates. Cell. 1996;87:9–12. doi: 10.1016/s0092-8674(00)81317-3. [DOI] [PubMed] [Google Scholar]
- Meyer E, Garnier O. Non-Mendelian inheritance and homology-dependent effects in ciliates. Adv Genet. 2002;46:305–337. doi: 10.1016/s0065-2660(02)46011-7. [DOI] [PubMed] [Google Scholar]
- Nowacki M, Haye JE, Fang W, Vijayan V, Landweber LF. RNA-mediated epigenetic regulation of DNA copy number. Proc Natl Acad Sci U S A. 2010;107:22140–22144. doi: 10.1073/pnas.1012236107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Landweber LF. Epigenetic inheritance in ciliates. Current Opinion in Microbiology. 2009;12:638–643. doi: 10.1016/j.mib.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Shetty K, Landweber LF. RNA-Mediated Epigenetic Programming of Genome Rearrangements. Annu Rev Genom Hum G. 2011;12:367–389. doi: 10.1146/annurev-genom-082410-101420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–154. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E. Mapping the germ-line and somatic genomes of a ciliated protozoan, Tetrahymena thermophila. Genome Research. 1998;8:91–99. doi: 10.1101/gr.8.2.91. [DOI] [PubMed] [Google Scholar]
- Phadke SS, Zufall RA. Rapid diversification of mating systems in ciliates. Biol J Linn Soc. 2009;98:187–197. [Google Scholar]
- Preer JR. Surface antigens of Paramecium. In: Gall J, editor. Molecular Biology of the Ciliated Protozoa. Academic Press; London: 1986. pp. 301–339. [Google Scholar]
- Przybos E, Tarcz S. Paramecium jenningsi complex: existence of three cryptic species confirmed by multi-locus analysis and strain crosses. Syst Biodivers. 2016;14:140–154. [Google Scholar]
- Raikov IB. The Protozoan Nucleus: Morphology and Evolution. Springer-Verlag; Wien: 1982. [Google Scholar]
- Schoeberl UE, Kurth HM, Noto T, Mochizuki K. Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Gene Dev. 2012;26:1729–1742. doi: 10.1101/gad.196493.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeberl UE, Mochizuki K. Keeping the Soma Free of Transposons: Programmed DNA Elimination in Ciliates. Journal of Biological Chemistry. 2011;286:37045–37052. doi: 10.1074/jbc.R111.276964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MC, Kusch J. Communicative functions of GPI-anchored surface proteins in unicellular eukaryotes. Crit Rev Microbiol. 2013;39:70–78. doi: 10.3109/1040841X.2012.691459. [DOI] [PubMed] [Google Scholar]
- Simon MC, Schmidt HJ. Antigenic variation in ciliates: Antigen structure, function, expression. Journal of Eukaryotic Microbiology. 2007;54:1–7. doi: 10.1111/j.1550-7408.2006.00226.x. [DOI] [PubMed] [Google Scholar]
- Singh DP, Saudemont B, Guglielmi G, Arnaiz O, Gout JF, Prajer M, Potekhin A, Przybos E, Aubusson-Fleury A, Bhullar S, Bouhouche K, Lhuillier-Akakpo M, Tanty V, Blugeon C, Alberti A, Labadie K, Aury JM, Sperling L, Duharcourt S, Meyer E. Genome-defence small RNAs exapted for epigenetic mating-type inheritance. Nature. 2014;509:447– 452. doi: 10.1038/nature13318. [DOI] [PubMed] [Google Scholar]
- Sonneborn TM. Acquired immunity to specific antibodies and its inheritance in P. aurelia. Proc Indiana Acad Sci. 1943;52:190–191. [Google Scholar]
- Sonneborn TM. Methods in Paramecium research. Methods Cell Physiol. 1970;4:241–339. [Google Scholar]
- Sonneborn TM. Paramecium-Aurelia Complex of 14 Sibling Species. T Am Microsc Soc. 1975;94:155–178. [Google Scholar]
- Sonneborn TM. Genetics of cellular differentiation - stable nuclear differentiation in eukaryotic unicells. Annual Review of Genetics. 1977;11:349–367. doi: 10.1146/annurev.ge.11.120177.002025. [DOI] [PubMed] [Google Scholar]
- Swart EC, Nowacki M. The eukaryotic way to defend and edit genomes by sRNA-targeted DNA deletion. Ann Ny Acad Sci. 2015;1341:106–114. doi: 10.1111/nyas.12636. [DOI] [PubMed] [Google Scholar]
- Swart EC, Wilkes CD, Sandoval PY, Arambasic M, Sperling L, Nowacki M. Genome-wide analysis of genetic and epigenetic control of programmed DNA deletion. Nucleic Acids Research. 2014;42:8970–8983. doi: 10.1093/nar/gku619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The Epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- Xu K, Doak TG, Lipps HJ, Wang JM, Swart EC, Chang WJ. Copy number variations of 11 macronuclear chromosomes and their gene expression in Oxytricha trifallax. Gene. 2012;505:75–80. doi: 10.1016/j.gene.2012.05.045. [DOI] [PubMed] [Google Scholar]
- Yao MC, Duharcourt S, Chalker DL. Genome-wide rearrangements of DNA in ciliates. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. ASM Press; Washington D.C.: 2002. pp. 730–758. [Google Scholar]
- Zufall RA, McGrath CL, Muse SV, Katz LA. Genome architecture drives protein evolution in ciliates. Molecular Biology and Evolution. 2006;23:1681–1687. doi: 10.1093/molbev/msl032. [DOI] [PubMed] [Google Scholar]