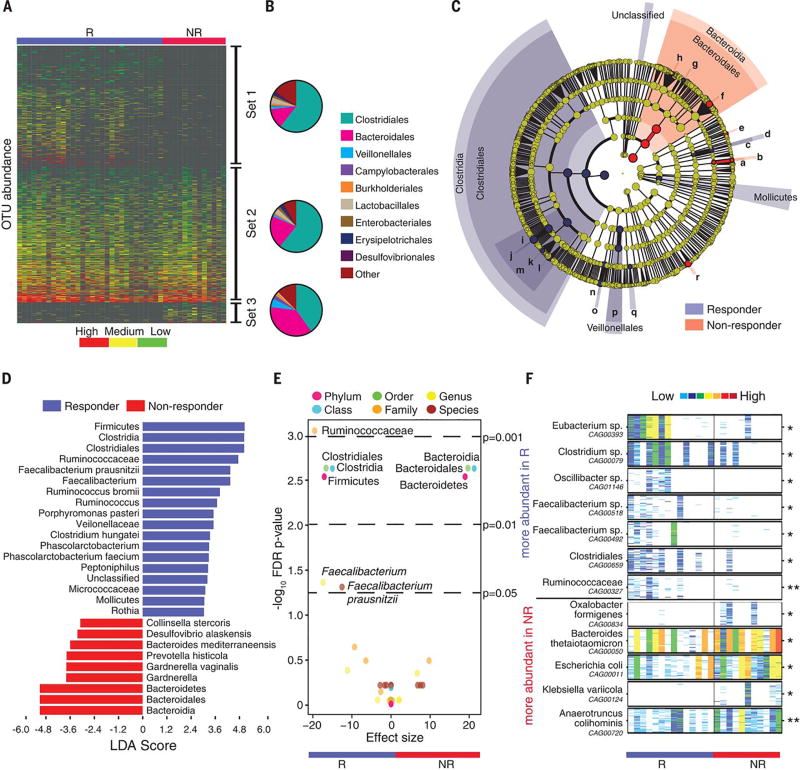
Figure 2. Compositional differences in the gut microbiome are associated with responses to anti-PD-1 immunotherapy.
(A) Heatmap of OTU abundances in R (n=30) and NR (n=13). Columns denote patients grouped by response and sorted by diversity within R and NR groups; rows denote bacterial OTUs grouped into 3 sets according to their enrichment/depletion in R versus NR: Set 1 (enriched in R), Set 2 (unenriched), and Set 3 (enriched in NR), and then sorted by mean abundance within each set. (B) Phylogenetic composition of OTUs within each set at the order level. Set 1 (enriched in R); Set 2 (unenriched); Set 3 (enriched in NR). (C) Taxonomic cladogram from LEfSe showing differences in fecal taxa. Dot size is proportional to the abundance of the taxon. Letters correspond to the following taxa: (a) Gardnerella vaginalis, (b) Gardnerella, (c) Rothia, (d) Micrococcaceae, (e) Collinsella stercoris, (f) Bacteroides mediterraneensis, (g) Porphyromonas pasteri, (h) Prevotella histicola, (i) Faecalibacterium prausnitzii, (j) Faecalibacterium, (k) Clostridium hungatei, (l) Ruminococcus bromii, (m) Ruminococcaceae, (n) Phascolarctobacterium faecium, (o) Phascolarctobacterium, (p) Veilonellaceae, (q) Peptoniphilus, (r) Desulfovbrio alaskensis. (D) LDA scores computed for differentially-abundant taxa in the fecal microbiomes of R (blue) and NR (red). Length indicates effect size associated with a taxon. p=0.05 for the Kruskal-Wallis test; LDA score > 3. (E) Differentially-abundant gut bacteria in R (blue) vs NR (red) by MW test (FDR-adjusted) within all taxonomic levels. (F) Pairwise comparisons by MW test of abundances of metagenomic species (MGS) identified by metagenomic WGS in fecal samples (n=25): R (n=14, blue), NR (n=11, red). *p<0.05, **p<0.01. Colors reflect gene abundances visualized using “barcodes” with the following order of intensity: white(0)<light blue<blue<green<yellow<orange<red for increasing abundance and each color change corresponds to a 4x fold abundance change. In these barcodes, MGS appear as vertical lines (co-abundant genes in a sample) colored according to the gene abundance.