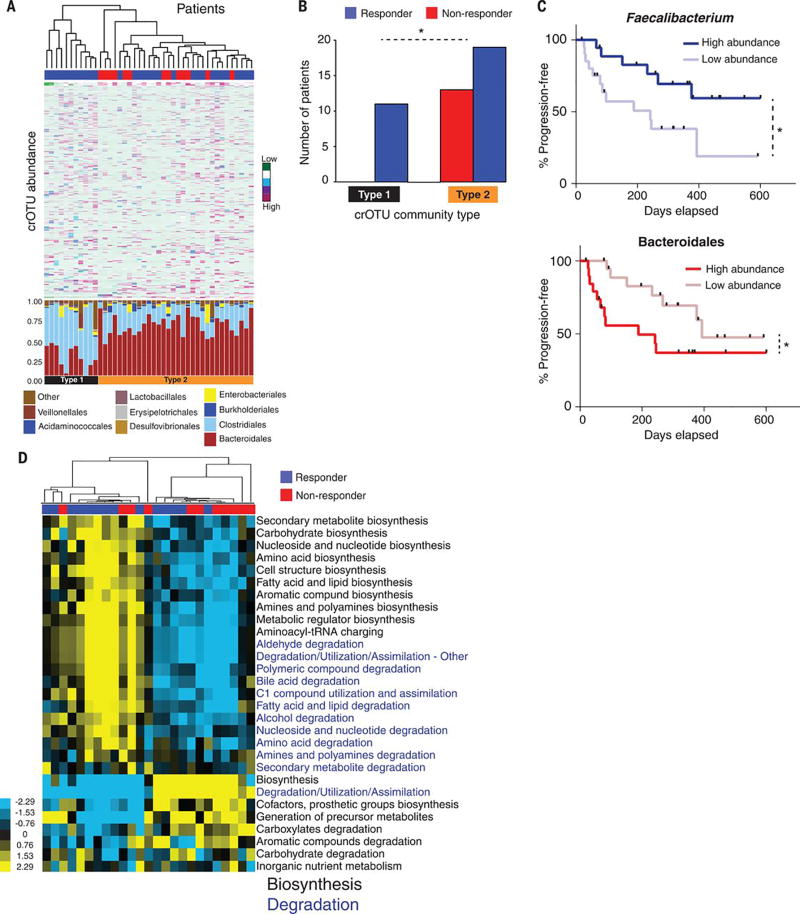
Figure 3. Abundance of crOTUs within the gut microbiome is predictive of response to anti-PD-1 immunotherapy.
(A) Top: Unsupervised hierarchical clustering by complete linkage of Euclidean distances of crOTU abundances in 43 fecal samples. Bottom: Stacked bar plot of relative abundances at the order level by crOTU community-type. (B) Association of crOTU community types with response to anti-PD-1 by Fisher’s exact test. crOTU community type 1 (black, n=11: R=11, NR=0); crOTU community type 2 (orange, n=32: R=19, NR=13). Blue bars indicate responders, whereas red bars indicate non-responders. (C) Comparison KM PFS curves by long-rank test in patients with high abundance (dark blue, n=19, median PFS=undefined) or low abundance (light blue, n=20, median PFS=242 days) of Faecalibacterium (top PFS curve). High abundance (dark red, n=20, median PFS=188 days) or low abundance (light red, n=19, median PFS=393 days) of Bacteroidales (bottom PFS curve). (D) Unsupervised hierarchical clustering of pathway class enrichment calculated as the number of MetaCyc pathways predicted in the metagenomes of fecal samples from 25 patients (R=14, NR=11). Columns represent patient samples (blue=R, red=NR) and rows represent enrichment of predicted MetaCyc pathways (blue=low enrichment, black=medium enrichment, yellow= high enrichment). Black text: biosynthetic pathways, blue text: degradative pathways. *p<0.05.