Figure 5.
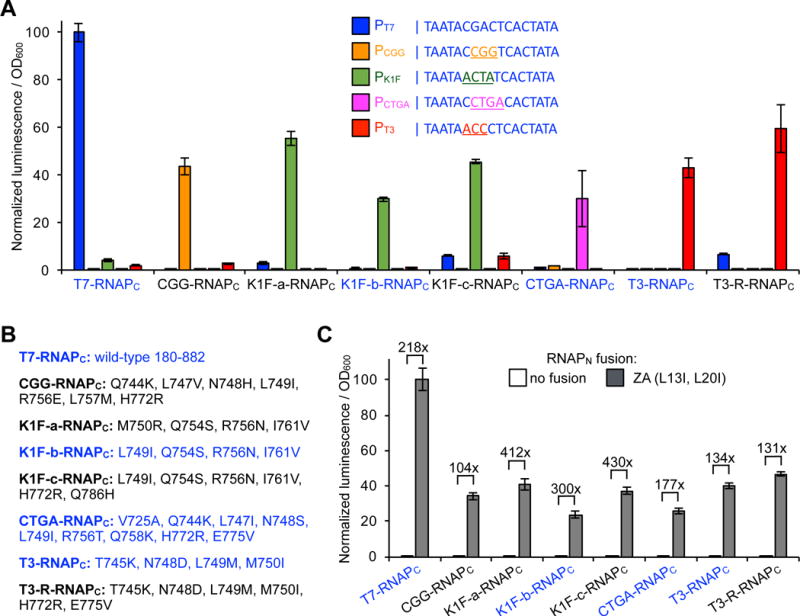
Uncovering a series of orthogonal proximity-dependent C-terminal split RNAPs. (A) E. coli transcriptional reporter assay for DNA promoter orthogonality of a series of split C-terminal T7 RNAP variants fused to ZB (ZB-RNAPC) on putative DNA promoter sequences. Sequences for the five target DNA promoters are shown (T7-blue, CGG-orange, K1F-green, CTGA-magenta, T3-red). E. coli transformed with the vector system shown in Figure 1B: 1) an expression vector for an evolved N-terminal RNAP fused to ZA (RNAPN-ZA), 2) an expression vector for a target RNAPC variant to be tested, and 3) a reporter vector that drives luciferase based on a target DNA promoter sequence. Cells were induced for 3 h with arabinose and then analyzed for luminescence. Error bars are ± s.e.m., n = 4. (B) Mutations shown for each RNAPC variant tested. (C) Proximity-dependent assembly of the series of ZB-RNAPC variants. E. coli transformed with the vector system shown in Figure 1B: 1) an expression vector for RNAPN fused to either nothing as a negative control or to ZA, 2) an expression vector for a target RNAPC variant to be tested, fused to ZB, and 3) a reporter vector that drives luciferase based on the target DNA promoter sequence of the variant being tested. Cells were induced for 3 h with arabinose and then analyzed for luminescence. Error bars are ± s.e.m., n = 4. The variants that were selected for further study are color-coded in blue.
