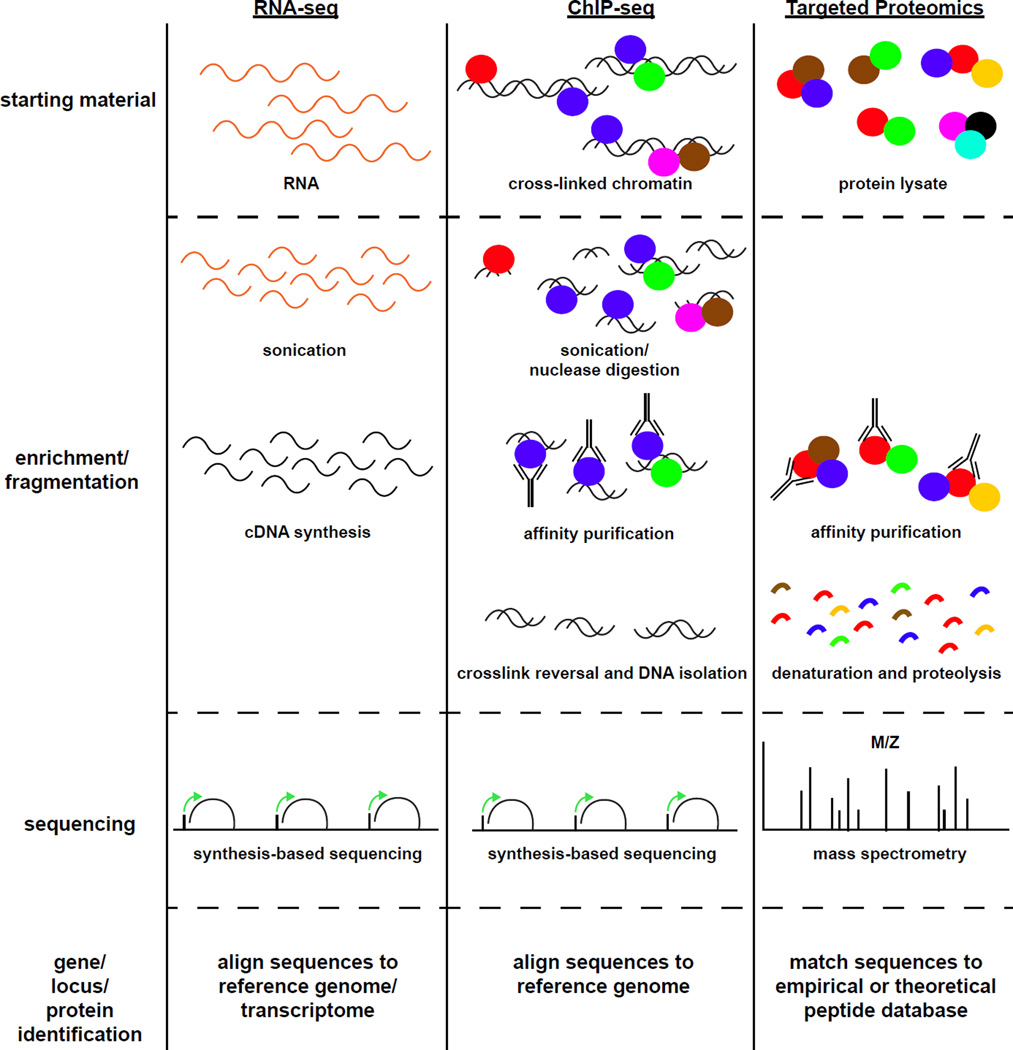
Figure 1. An overview of large-dataset experimental approaches.
RNA-seq, ChIP-seq, and proteomic analyses follow similar workflows. Briefly, starting material to be analyzed is isolated from cells or tissue. Enrichment steps, such as affinity purifications, can be employed to narrow the population of input material analyzed. Molecules of interest are then fragmented, either mechanically or enzymatically, to permit massively parallel sequence analysis. Resulting sequence data is compared to a reference database (either a transcriptome, a genome, or a peptide sequence collection) to attain identities of the molecules of interest.