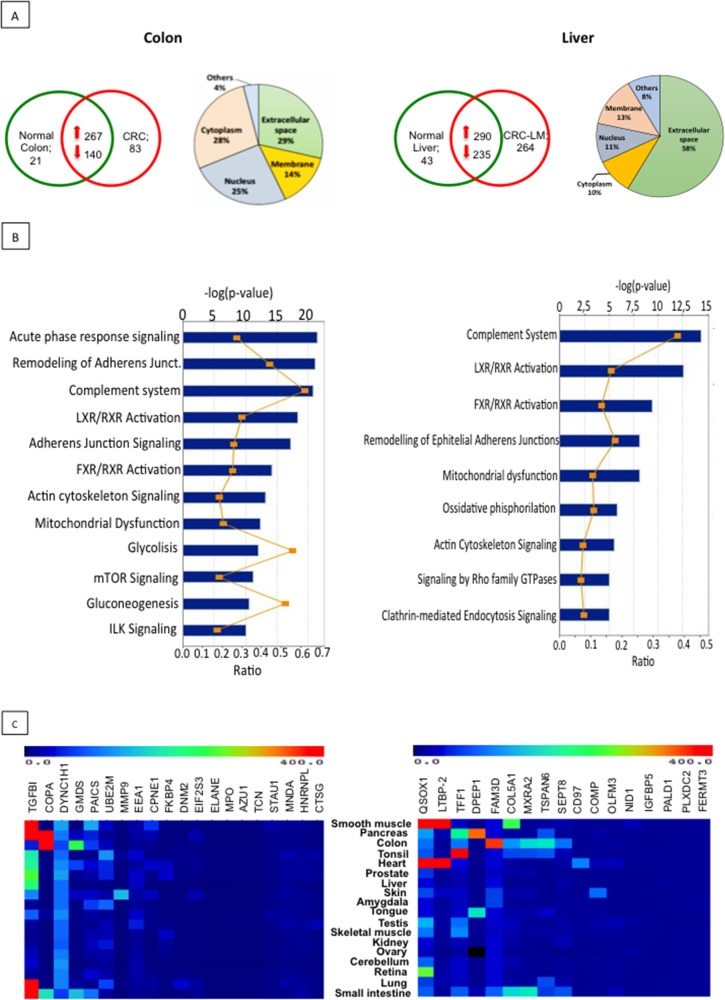
Figure 2. Proteomic analysis of EXPEL extruded fluid identifies potential cancer biomarkers.
(A) Absolute numbers of proteins, identified after proteomic analysis of EXPEL extruded fluids, in at least 4 out of 7 for CRC (upper, left panel) and 3 out of 6 for CRC-LM (upper right, panel) replicates. Pie charts indicating the predicted sub-cellular localization of identified proteins for CRC (on left side) and CRC-LM (on right side) are shown. (B) Significantly altered canonical pathways of cancer up-regulated and uniquely expressed proteins analyzed by the IPA software using IPA Core Analysis for colon (on the left) and liver (on the right) EXPEL extruded fluids. The canonical pathways are shown along the y-axis of the bar chart. The x-axis indicates the statistical significance (on the upper part, calculated using the right-tailed Fisher exact test. The P value indicates which biologic annotations are significantly associated with the input molecules relative to all functionally characterized mammalian molecules. “Ratio” (differential yellow line and markers) refers to the number of molecules from the dataset that map to the pathway listed divided by the total number of molecules that map to the canonical pathway from within the IPA knowledgebase. (C) Normal tissue gene expression of potential biomarkers candidates discovered from colorectal cancer (left panel) and liver metastases (right panel). mRNA expressions were assessed using BioGPS public database.