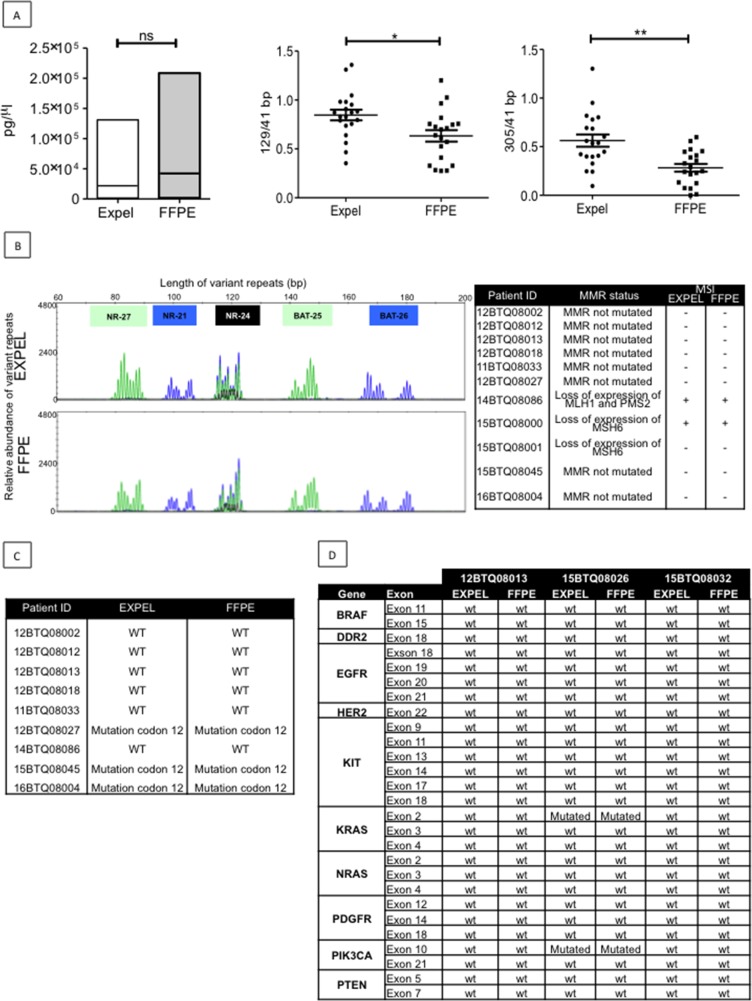
Figure 4. EXPEL extruded fluid contains high quality tumor DNA (tDNA) that is exploitable for genetic profiling.
(A) Left panel, equivalent yield of tDNA was obtained from FFPE sections and matched EXPEL fluids of CRC primary lesions (n=10) and CRC-liver metastatic lesions (n=10). The quality of the DNA was assessed on the same extracts. The ratios of long 129bp (middle panel) and 305bp (right panel) to short amplified fragments (41bp) indicated that EXPEL tDNA presented with significantly higher amounts of long fragments in comparison with the FFPE tDNA. Dot plots show the mean ± SEM. Statistical significance was calculated using Wilcoxon paired test (* p<0.05, ** p<0.01 and *** p<0.001). (B) PCR microsatellite instability (MSI) analysis at 5 loci (NR-27, NR-21, NR- 24, BAT-25 and BAT-26) of a representative CRC primary tumor shows similar electropherograms for both FFPE and EXPEL tDNA extracts (left panel). Right panel summarizes MSI analysis results obtained for a subset of 11 tumor samples. The corresponding MMR status, based on routine IHC detection of MMR genes (MLH1, MSH2, MSH6, PMS2), is given. (C) Concordant KRAS mutational status (codon 12) detected using pyrosequencing on FFPE and EXPEL tDNAs isolated from CRC primary lesions and CRC-liver metastatic lesions. (D) Next-generation sequencing (NGS) technique used for the detection of 10 cancer related genetic alterations showed identical results on FFPE and EXPEL tDNAs isolated from CRC and CRC-LM lesions.