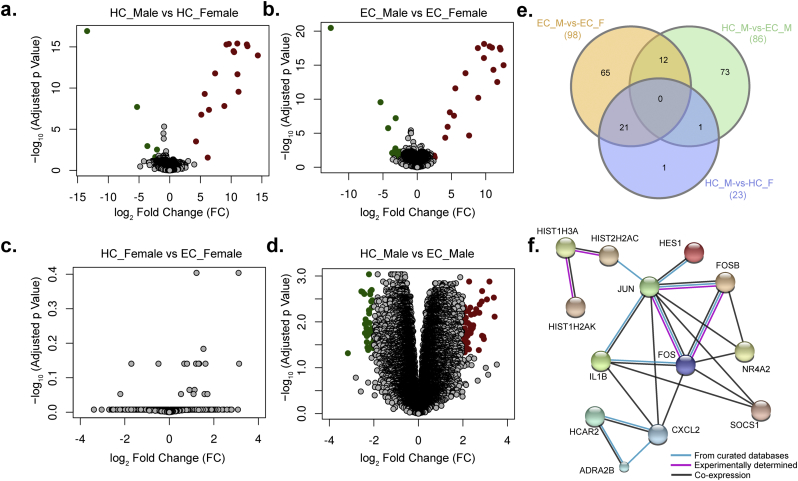
Fig. 3.
Differential expression (DE) of the transcript (protein-coding and non-protein coding) expression profile in a gender-specific manner. (a–d) DE are represented as volcano plots in different categories of the HC and EC groups were categorized into males (HC_M: n = 7; EC_M: n = 10) and females (HC_F: n = 6; EC_F: n = 9) before analysis. Transcripts with log2-fold-change value greater than two and adjusted p-value (FDR) < 0.05 (filled red circle), were considered as upregulated and those with less than minus two and adjusted p-value (FDR) < 0.05 (filled green circle) were considered as downregulated. (e) Venn diagram of DE transcripts. The sum of the numbers in each large circle represents the total number of differentially expressed genes among various combinations (EC_M vs. EC_F, HC_M vs. EC_M and HC_M vs. HC_F). The overlapping part of the circles represents common DEGs between combinations. (f) The network analysis in STRING with 38 proteins that were upregulated in female EC compared to the male EC.