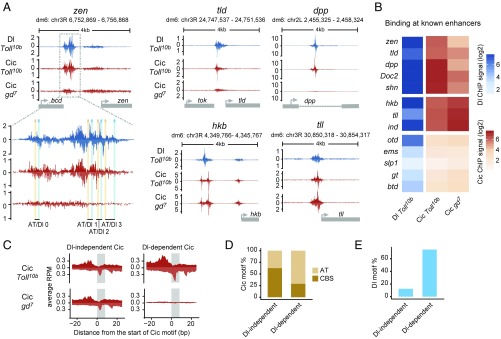
Fig. 3.
Dorsal mediates binding of Cic to low-affinity AT sites. (A) ChIP-nexus profiles of Dl and Cic at the indicated genes in Toll10b or gd7 mutant background; a zoom-in view of the zen VRE enhancer is included. Peaks on the upper (positive) and lower (negative) strands represent pile-ups of 5′ read ends after digestion with exonuclease, which stops at sites of protein–DNA cross-linking. Cic binding to zen, tld, and dpp (but not to hkb and tll) is strongly reduced in gd7 embryos lacking nuclear Dorsal protein. (B) Heat maps of Dorsal and Cic binding at known enhancers. ChIP-nexus signal was calculated within a 200-bp region centered on the highest Dorsal summit. (C) Average ChIP-nexus signals for Dorsal-independent (n = 56) and Dorsal-dependent (n = 59) Cic binding motifs across the genome. RPM, reads per million. (D) Type of Cic motif found in the Dorsal-independent and Dorsal-dependent sets: CBS motifs are an exact match to TSAATGAA, while AT motifs contain one mismatch relative to this consensus site. (E) Fraction of Cic binding motifs flanked by Dorsal binding motifs (GGRWWTTCC with up to one mismatch) within less than 50 bp.