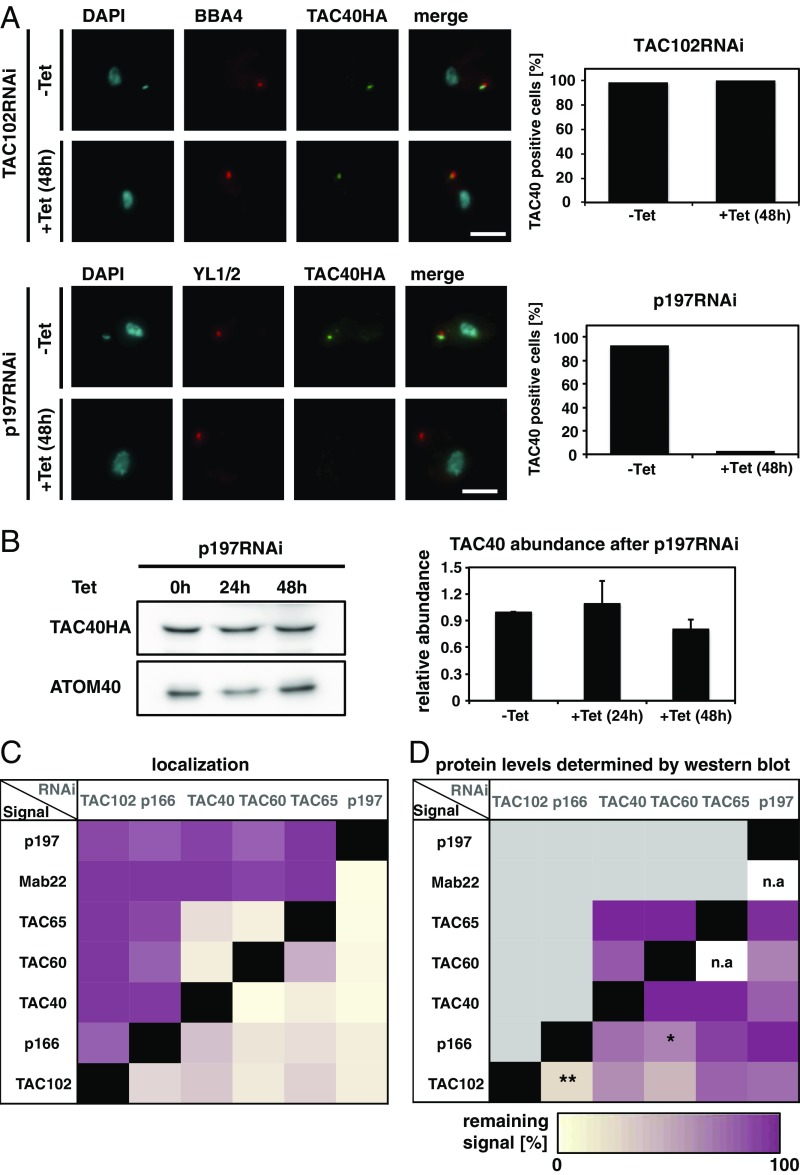
Fig. 2.
Dependence of TAC assembly on the individual components. (A) Fluorescence microscopy images of TAC102 RNAi or p197 RNAi uninduced (−Tet) or 48 h-induced (+Tet) cells, stained with DAPI (DNA, cyan) and probed with antibodies BBA4 or YL1/2 (basal body, red) and anti-HA (TAC40HA, green). Next to it, the quantification of TAC40-positive cells [cells without a YL1/2 signal were excluded (n ∼ 100)]. (B) Protein abundance of TAC40 determined by Western blotting from cells uninduced and 24 h and 48 h post-p197 RNAi induction. Bar graph shows protein abundance (n = 3). ATOM40 abundance was used as a loading control. (C) Quantification of each protein as described in A. Purple indicates that in 100% of the cells, the signal can correctly localize, whereas 0% is indicated by yellow. A black box shows that a knockdown of this component results in a loss of the signal for the same protein. (D) Quantification of each protein as described in B. The percentage of the remaining signal after inducing for 48 h in comparison with the uninduced cells was calculated. Yellow indicates loss of signal in Western blotting, while purple indicates no changes. A black box shows that a knockdown of this component results in a loss of the signal for the same protein, and a gray box implies no investigation on Western blot since no changes occurred in localization; n/a, not applicable. The P value was calculated by two-tailed heteroscedastic t test to perform significance measurements. *P ≤ 0.05; **P ≤ 0.01. (Scale bar: 3 µm.)