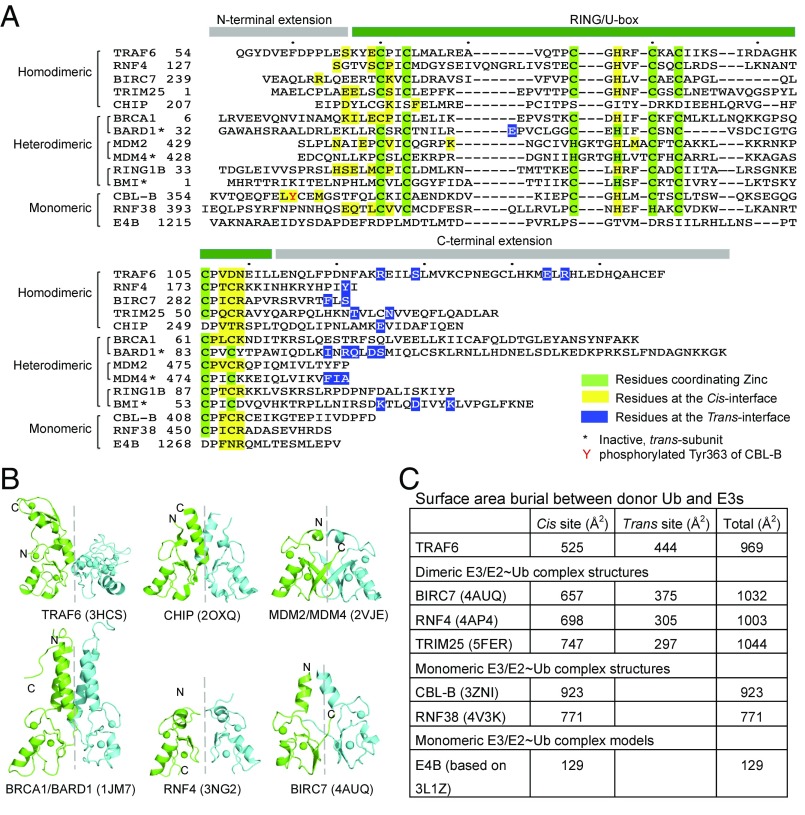
Fig. 5.
Analysis of diverse RING/U-box ubiquitin ligases. (A) Sequence alignment of representative RING/U-box E3s. Residues of RING/U-box E3s involved in donor ubiquitin engagement were mapped either from real ternary complex structures or from modeled complex structures. Residues at the cis-interface are colored in yellow, while those at the trans-interface are colored in blue. Zinc-coordinating residues are marked in green. (B) Cartoon diagrams of TRAF6 RING dimer (PDB ID: 3HCS), CHIP U-box dimer (PDB ID: 2OXQ), MDM2/MDM4 RING dimer (PDB ID: 2VJE), BRCA1/BARD1 RING dimer (PDB ID: 1JM7), RNF4 RING dimer (PDB ID: 3NG2), and BIRC7 RING dimer (PDB ID: 4AUQ). When the RING/U-box protomers colored in green are superimposed, the symmetric RING protomers colored in cyan do not show similar orientations, indicating differences in RING/U-box dimer architectures. (C) Buried surface areas between donor-Ub and diverse RING/U-box E3 ubiquitin ligases. The buried interfaces were calculated by PDBePISA using crystal structures of BIRC7/UbcH5b∼Ub (PDB ID: 4AUQ), RNF4/UbcH5a∼Ub (PDB ID: 4AP4), TRIM25/UbcH5a∼Ub (PDB ID: 5FER), CBL-B/UbcH5b∼Ub (PDB ID: 3ZNI), and RNF38/UbcH5b∼Ub (PDB ID: 4V3K) and structural models of TRAF6/Ubc13∼Ub and E4B/UbcH5c∼Ub.