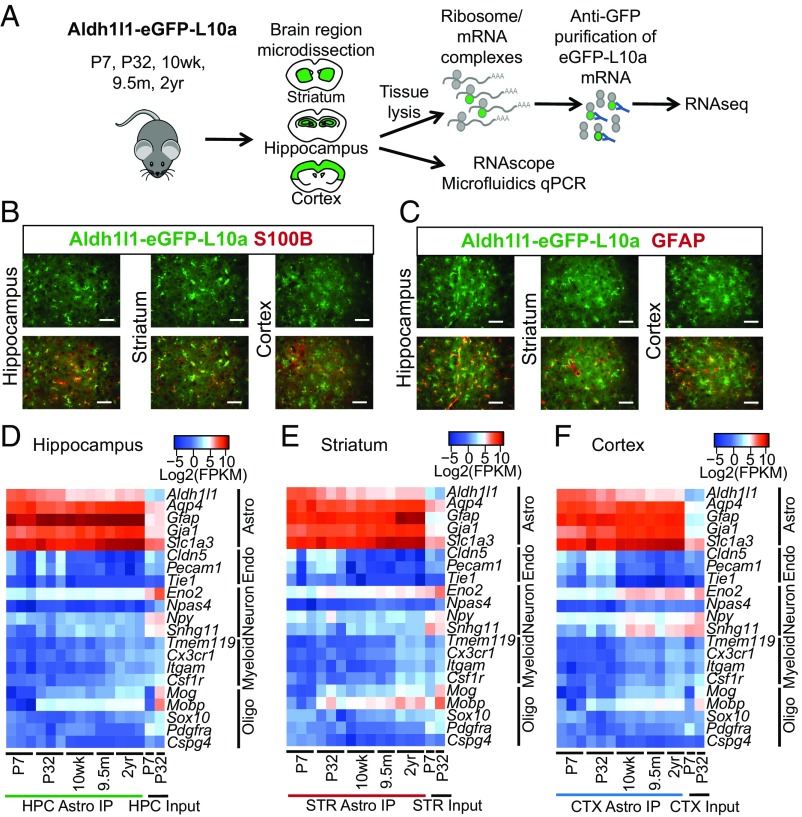
Fig. 1.
Isolation and validation of the aging astrocyte transcriptome from distinct brain regions. (A) Schematic diagram showing the experimental strategy for isolation of astrocyte RNA by TRAP across the lifespan of the mouse, and validation of differentially expressed aging genes. (B and C) Representative images of astrocytes from the hippocampus, striatum, and cortex in an adult (10 wk old) Aldh1l1-eGFP-L10a mouse showing costaining for (B) S100β and (C) GFAP. (Scale bar, 50 µm.) (D–F) Cell purity heatmaps depicting mean FPKM expression values for cell-type−specific markers of astrocytes, neurons, oligodendrocyte lineage cells (oligo), endothelial cells (endo), and microglia/macrophages (myeloid) in (D) hippocampal, (E) striatal, and (F) cortical astrocyte and input samples. CTX, cortex; HPC, hippocampus; IP, immunoprecipitation; STR, striatum. See also Datasets S1–S4.