Figure 2.
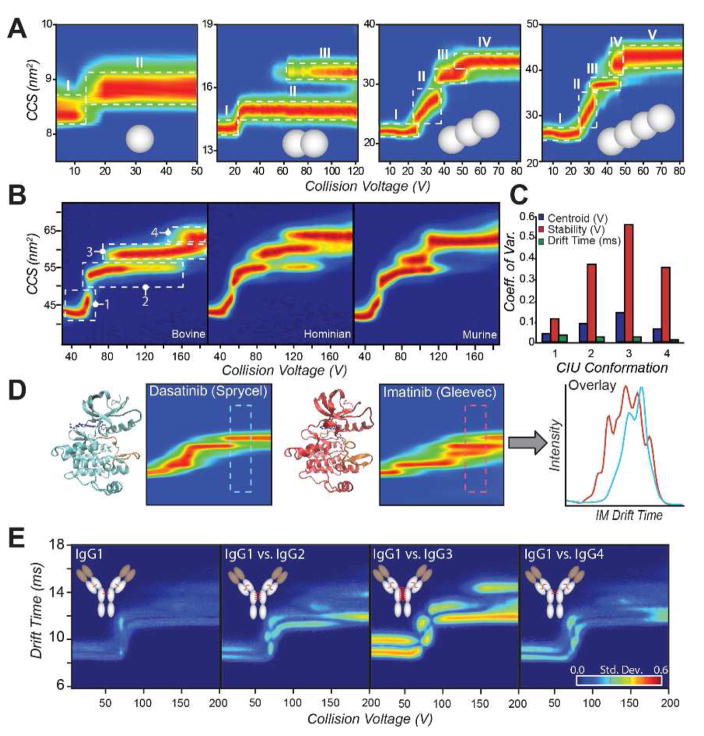
A: A series of covalently linked poly-ubiquitin proteins (1–4 ubiquitins, gray spheres) is probed by CIU [37]**. Single domain ubiquitin results in a single CIU transition, from an initial native-like state (I) to a more extended state (II) upon collisional activation. Each additional domain added results in an additional CIU transition, indicating that the transitions are representative of the domain structure of the protein in solution. B: Bovine, human, and murine serum albumin proteins CIU fingerprints are compared. Despite high sequence homology and globally similar three-dimensional structures, CIU readily distinguishes each variant, demonstrating sensitivity towards subtle alterations in protein isoforms [13]**. C: Coefficient of variation (CV) across the bovine, human, and murine albumins represented in (B) for centroid voltage (blue), stability or horizontal length (red), and center drift time (green) for each feature. High CVs indicate significant differences between fingerprints. D: Comparison of type I (Dasantinib, left) and type II (Imatinib, right) inhibitors bound to Abl kinase. CIU distinguishes the binding location of inhibitors to the kinase, enabling a screening assay based on the region of maximal difference in the CIU fingerprint (far right) [17]. E: IgG subtypes 1–4 (left to right) are quantitatively distinguished by CIU [45]**. Each subtype exhibits different patterns of disulfide bonding in a broadly conserved overall structure, resulting in different CIU fingerprints.