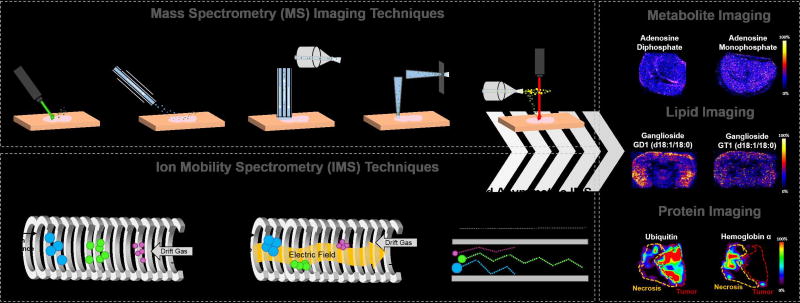
Figure 4.
Examples of peptide and protein imaging from biological tissue samples using MS imaging coupled to IMS. The left panel shows the application of MALDI and TWIMS in the “IMS-TAG” method, which employs an internal standard protein that, when digested, yields peptides of interest (denoted with red arrows) within a tissue sample. The images show the distribution of three peptides within the ion images, a vimentin peptide, a HS90 peptide, and a histone H2A peptide, where only vimentin and HS90 are within the tissue [35]. The middle panel shows the use of LESA and FAIMS to image proteins from mouse brain tissue. An increase in the S/N and number of proteins detected with LESA was observed when FAIMS was used. The spectra when FAIMS voltage are applied shows greater variation in the species within a mouse brain tissue section observed within the same analysis time, and the images show a higher intensity of the calmodulin protein when FAIMS is used. Similarly, left panel shows the use of the LMJ-SS and FAIMS to image proteins in rat brain tissue. Increased ion intensity of protein species from rat brain tissue and improved spatial correlation of the molecular ions within the tissue section are observed, with the thymosin β-4 located majorly in the hippocampus of the rat brain [56].