Abstract
Anthracyclines have been a cornerstone in the cure of diffuse large B-cell lymphoma (DLBCL) and other hematological cancers. The ability of anthracyclines to eliminate DLBCL depends on the presence of topoisomerase-II-alpha (TopIIA), a DNA repair enzyme complex. We identified nucleolin as a novel binding partner of TopIIA. Abrogation of nucleolin sensitized DLBCL cells to TopIIA targeting agents (doxorubicin/etoposide). Silencing nucleolin and challenging DLBCL cells with doxorubicin enhanced the phosphorylation of H2AX (γH2AX-marker of DNA damage) and allowed DNA fragmentation. Reconstitution of nucleolin expression in nucleolin-knockdown DLBCL cells prevented TopIIA targeting agent-induced apoptosis. Nucleolin binding to TopIIA was mapped to RNA binding domain 3 of nucleolin, and this interaction was essential for blocking DNA damage and apoptosis. Nucleolin silencing decreased TopIIA decatenation activity, but enhanced formation of TopIIA-DNA-cleavable complexes in the presence of etoposide. Moreover, combining nucleolin inhibitors: aptamer AS1411 or nucant N6L with doxorubicin reduced DLBCL cell survival. These findings are of clinical importance because low nucleolin levels versus high nucleolin levels in DLBCL predicted 90 month estimated survival of 70% versus 12% (P<0.0001) of patients treated with R-CHOP based therapy.
INTRODUCTION
Diffuse large B-cell lymphoma (DLBCL) is the most common histology of non-Hodgkin lymphoma (NHL) and has a variable response to anthracycline-containing regimens (ACRs).1 Despite the recent improvement in therapy with the addition of immunotherapy to ACRs, only 50% of DLBCL patients are cured.2 In our search for improving DLBCL outcomes, we examined cell response to therapy.
Topoisomerase II alpha (TopIIA) is a multifunctional nuclear enzyme required during DNA replication, transcription, chromosome segregation, and DNA damage repair because of its ability to relax supercoiled DNA and decatenate/catenate double-stranded DNA rings. TopIIA also plays an important role in cell proliferation and chemotherapy resistance.3, 4 TopIIA targeting agents doxorubicin and etoposide suppress the religation steps catalyzed by TopIIA, thus allowing accumulation of covalent TopIIA-DNA cleavage complexes.4 In breast cancer, the appearance of TopIIA gene amplification predicts a better response to anthracycline-based chemotherapy as well a longer overall survival in most studies.5–7
DNA double strand breaks (DSBs) arise through both endogenous (oxidative stress) and exogenous (ionization radiation or DNA damaging agents such as doxorubicin) insults inducing genomic instability. In response to the appearance of DSBs, cells undergo arrest of the cell cycle and initiate DNA repair, or if DNA damage is extensive, trigger apoptosis. In order to repair sites of DNA damage, both histone modification and chromatin remodeling are required to allow access for repair factors. Interestingly, nucleolin, which is normally a nucleolus-predominant protein, is drawn to DSBs induced by endonuclease or ɣ-radiation.8, 9 Nucleolin is expressed ubiquitously, but elevated expression is primarily noted in cancers including DLBCL and proliferation-associated cells such as cancer-associated endothelial cells.10–14 Nucleolin binds to several key DNA repair and recombination proteins such as topoisomerase-I, RAD50, MDC1, PCNA and γH2AX.8, 9, 15, 16 To date, the functional importance of nucleolin in DNA damage response and interactions with repair proteins are not completely understood. In this current study, we identified nucleolin binding to TopIIA and this interaction is essential for regulating TopIIA targeting agents induced DLBCL cell death. Further, we demonstrate that low levels of nucleolin predict better survival of patients with DLBCL.
MATERIALS AND METHODS
Cell culture
DLBCL (SU-DHL-2, SU-DHL-4, SU-DHL-6, HT) and Burkitt lymphoma (BJAB) cell lines were obtained from the American Type Culture Collection (ATCC). SU-DHL-9 DLBCL cells were a gift from Dr. Linda Baum (David Geffen School of Medicine, University of California, Los Angeles). All cells were maintained as per the vendor’s instruction. Cell lines were regularly tested for mycoplasma using MycoAlert and identity confirmed by STR analysis. The CD19-positive B cells were derived from peripheral blood B cells from healthy donors obtained from the Gulf Coast Blood Center (Houston, TX) and isolated using CD19-positive magnetic beads as described previously and stored.12 Patient DLBCL samples were collected with written consent of the patients at The University of Texas MD Anderson Cancer Center under research protocols LAB08-0190, 2008-0075, and 2005-0656 according to the Declaration of Helsinki. Primary DLBCL specimens having over 90% tumor cells to be included in the analysis after collection from effusions and lymphoma tumors. For protein expression, frozen stored DLBCL tissues and CD19 lymphocytes were thawed and processed side by side. Cells and tumor samples were lysed in cell lysis buffer (Cell Signaling Technology) supplemented with protease inhibitors (Roche) and calyculin A (Cell Signaling Technology) on ice, then combined with Laemmli sample buffer. Lysates were boiled and centrifuged in 4°C for 10 min at 10,000g followed by Western blotting.
Cell transfection and stable cell line generation
We knocked down nucleolin (NCL) in DLBCL cell lines by electroporation of specific nucleolin-targeting siRNA (AM16708; 144015 target exon 3; siR-1), control non-targeting (AM4635) ThermoFisher, SMARTpool-designed ON-TARGETplus siRNA (siR-2; J-003854-07, target exon 6) and siCONTROL non-targeting siRNA (siR-CON; D-001810) (Dharmacon/Thermo Scientific) using the Neon Transfection System according to the manufacturer’s instructions (Life Technologies). Stable nucleolin knockdown cells were generated using lentiviruses expressing human nucleolin shRNA (sh-NCL-2, Sigma; TRCN0000062283) targeting the UTR of nucleolin, cloned in pLKO.1 vector.17 Transduced cells were selected with puromycin (1μg/mL; Sigma-Aldrich). To reconstitute nucleolin expression in stable nucleolin-knockdown cells, plasmid (pCMV vector; Origene) encoding C-terminal FLAG (DDK)-tagged full-length or deleted domain constructs of nucleolin were transfected into the cells using electroporation and selected with neomycin (G418, 1.0 mg/mL; PAA Laboratories). Expression of exogenous nucleolin in the cells was confirmed with Western blotting.
Comet assay
DNA damage was measured using the comet assay.18 Briefly, cells were mixed with pre-warmed 0.75% ultra-low gelling agarose (44415 2G; BDH Electran, BDH Laboratory Supplies) and layered on cold microscopic slides precoated with 0.1% agarose. After incubation at 4°C, lysis was carried out using lysis buffer (2.5% sodium dodecyl sulfate, 1% sodium sarcosinate, and 25 mM ethylene-diaminetetraacetic acid, pH 9.5) for 15 minutes at 25°C to 30°C. Slides were washed for 5 minutes in distilled water at 10°C and electrophoresed (90 mM Tris base, 90 mM boric acid, 2.5 mM ethylene-diaminetetra-acetic acid, pH 8.3) at 2 V/cm for 5 minutes at 10°C. Cells were stained with propidium iodide and observed using fluorescent microscope. Randomly one hundred cells were scored for comet length from three independent experiments. The length of the comet was measured across all cells using the ImageJ software; statistical T-test was used to determine the significance of the experiment.
Immuno-histochemical Analysis
Expression of nucleolin and TopIIA proteins was performed on 104 DLBCL patients who were uniformly treated with R-CHOP regimen. Immunohistochemistry (IHC) analysis was performed on tissue microarrays (TMA) constructed with formalin-fixed, paraffin-embedded (FFPE) tissue using antibodies for Nucleolin (sc-55486; 1:6000) and TopIIA (12286; 1:600, Cell Signaling), as previously described.19–21 High versus low and positive versus negative cutoffs were determined based on survival analysis using the X-tile software (version 3.6.1, Yale School of Medicine, New Haven, CT). The nucleolin staining intensity and percentage of positive cells were analyzed independently by two hematopathologists (QY and KHY) and scored using the following grading system: staining intensity (0, absent; 1, low; 2, intermediate; 3, high); percentage of positive cells per every 5% increment. A nucleolin/TopIIA composite score was obtained from the sum of the scores for staining intensity and the percentage of positive cells (1, 0–1; 2, 2–3; 3, 4–5).
Statistical Analysis
Clinico-pathologic features and biomarker correlation were analyzed using the Fisher exact test. Overall survival (OS) and progression-free survival (PFS) Kaplan-Meier analyses were performed using the GraphPad Prism-6 (GraphPad Software, San Diego, CA). Data reported as means ± standard error of the mean for three independent experiments. Differences were compared between groups using the two-tailed Student’t-test. All differences with P ≤ 0.05 were considered statistically significant.
RESULTS
Nucleolin is overexpressed in DLBCL cells
Nucleolin protein expression was analyzed in DLBCL cell lines (SU-DHL-2,4,6,9 and HT), DLBCL tumors; BJAB cells (positive control);12 and normal B cells from healthy donors by Western blot analysis. DLBCL cell lines had higher levels of nucleolin expression over healthy donor B cells (Figure 1a). In addition, primary DLBCL tissues samples had high, but variable nucleolin expression (Figure 1b). To evaluate nucleolin expression in DLBCL, we performed immuno-histochemical (IHC) staining for nucleolin in 104 DLBCL patients. This patient population is representative of individuals presenting with DLBCL which were mostly elderly males with advanced stage DLBCL (Table S1). Expression of nucleolin was found in 58 (55.7%) of 104 DLBCL cases. Figure 1c shows representative nucleolin staining. There was no significant difference of nucleolin expression between GCB and ABC subtypes (P=0.058) (Data not shown). The immuno-histochemical cutoff for nucleolin overexpression associated with significant prognostic impact was determined by X tile approach with maximum specificity and sensitivity. Using this method, the cutoff for nucleolin expression (nucleolin high) was set at ≥50%; 58 (55.7%) of 104 patients had nucleolinhigh DLBCL. These patients had significantly worse OS (P<0.0001) and PFS (P<0.0001) than patients with nucleolin low DLBCL (Figure 1d)). The 5-year OS rate was ~35% for patients with nucleolinhigh DLBCL and ~80% for patients with nucleolinlow DLBCL.
Figure 1.
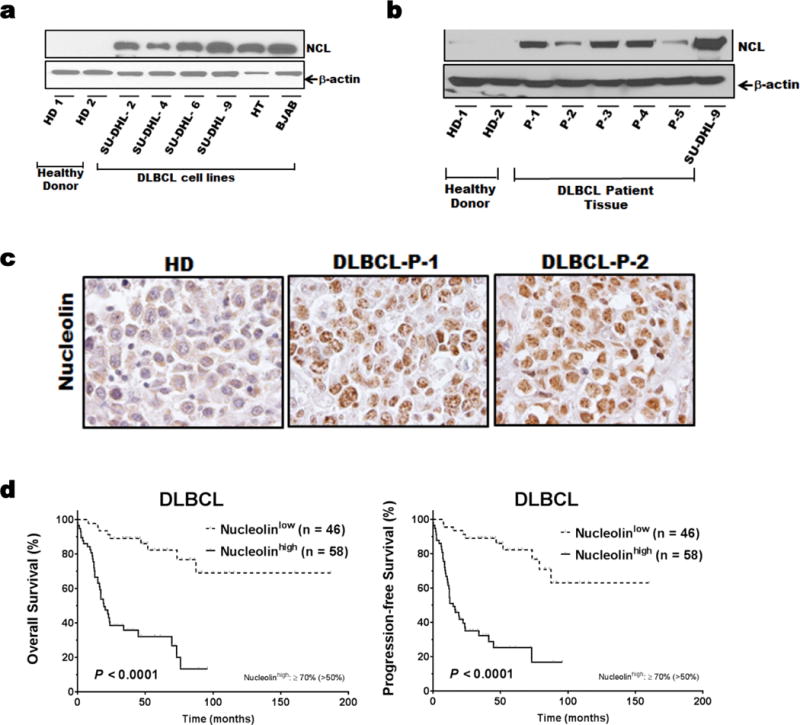
Nucleolin expression in DLBCL patient samples and cell lines.
Whole-cell lysates from the DLBCL cell lines SU-DHL-2,-4,-6,-9, and HT (a) primary DLBCL tissues and B-lymphocytes from healthy donors (b) were subjected to Western blot analysis for nucleolin and β-actin. (c) Representative Immunohistochemically staining of nucleolin in DLBCL patients and normal lymph node, 600X magnification. (d) Prognostic impact of nucleolin expression on overall survival (OS) and progression-free survival (PFS) in DLBCL (included on TMA) by Kaplan-Meier method.
To further investigated the robustness of our results obtained for nucleolin protein expression by IHC, we analyzed the clinical outcomes using RNA expression data sets (two different cohorts of 119 patients (GSE4475) and 414 patients (GSE10846)) with DLBCL treated with CHOP-based therapy.22, 23 Patients in each cohort were divided into two groups with high and low nucleolin mRNA levels.24 The high nucleolin RNA expressing groups in both patient cohorts had a significantly lower overall survival rate, as compared to the low nucleolin expressing groups (Supplementary Figure S1b). This is of fundamental interest as nucleolin is directly implicated in determining the fate of cells undergoing repair after induced DNA damage.8
Nucleolin inhibits DLBCL cell death induced by TopIIA targeting agents
Nucleolin has previously shown to localize to DNA repair sites. Thus we first analyzed the levels of doxorubicin/etoposide-induced apoptosis after silencing nucleolin expression with two independent siRNAs (Figure 2a). In all 3 DLBCL cell lines, the knockdown of nucleolin led to enhanced rates of TopIIA targeting agent-mediated cell apoptosis (Figure 2b and Supplementary Figure S2a). However, treatment with the TopIIA enzyme inhibitor dexrazoxane, failed to induce cell death, suggesting that catalytic activity is not required for TopIIA targeting agent-mediated cell death (data not shown). Moreover, doxorubicin treatment generated higher levels of cleavage products of caspase 3, PARP and reduced BID expression in nucleolin-silenced cells, as compared to control (Figure 2c). Overall, nucleolin suppressed TopIIA targeting agent-mediated apoptosis in DLBCL.
Figure 2.
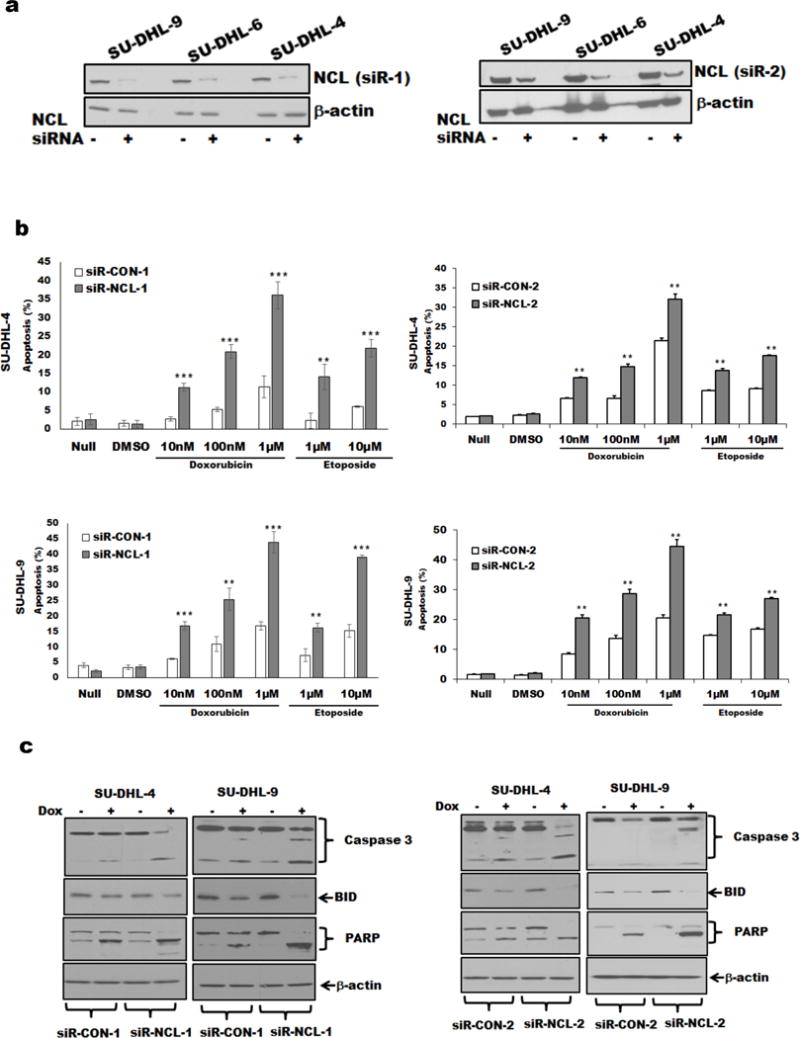
DLBCL cells with nucleolin knockdown and rates of apoptosis induced by TopIIA targeting agents.
Nucleolin suppresses TopIIA targeting agent-induced DNA damage in DLBCL cells
Nucleolin exerts a plethora of functions, so to clarify its role in response to treatment with TopIIA targeting agents,25 we examined DLBCL cells for DNA fragmentation as analyzed by the comet assay. Treatment with 0.1μM doxorubicin or 1μM etoposide caused comet-shaped distribution of DNA that was pronounced in nucleolin-knockdown cells (Figure 3a). Phosphorylation of histone H2AX (γH2AX) at Ser-139 is an accepted marker for DSBs.26 Treatment with doxorubicin or etoposide induced phosphorylation of H2AX in a dose and time dependent manner, while having no impact on the levels of TopIIA and H2AX (Figure 3b). Silencing nucleolin followed by treatment with doxorubicin enhanced the phosphorylation of nuclear H2AX, again supporting the notion that the loss of nucleolin combined with a TopIIA targeting agent favors DNA damage (Supplementary Figure S2b). In this study, the amount of drug used is sufficient to induce DNA damage leading to apoptosis or cell stress leading to DSB repair as reported previously.27,28,29 Integrity of DNA depends on a balance between the rate of DNA damage and DNA repair. We evaluated the contribution of nucleolin to DNA repair by reducing basal nucleolin expression by 70%. Concomitant treatment with NU7026, a DNA-dependent protein kinase inhibitor and doxorubicin,30 increased the γH2AX levels in nucleolin-silenced DLBCL cells as compared to control cells, implying that DNA repair was aided by the presence of NU7026 (Figure 3c).
Figure 3.
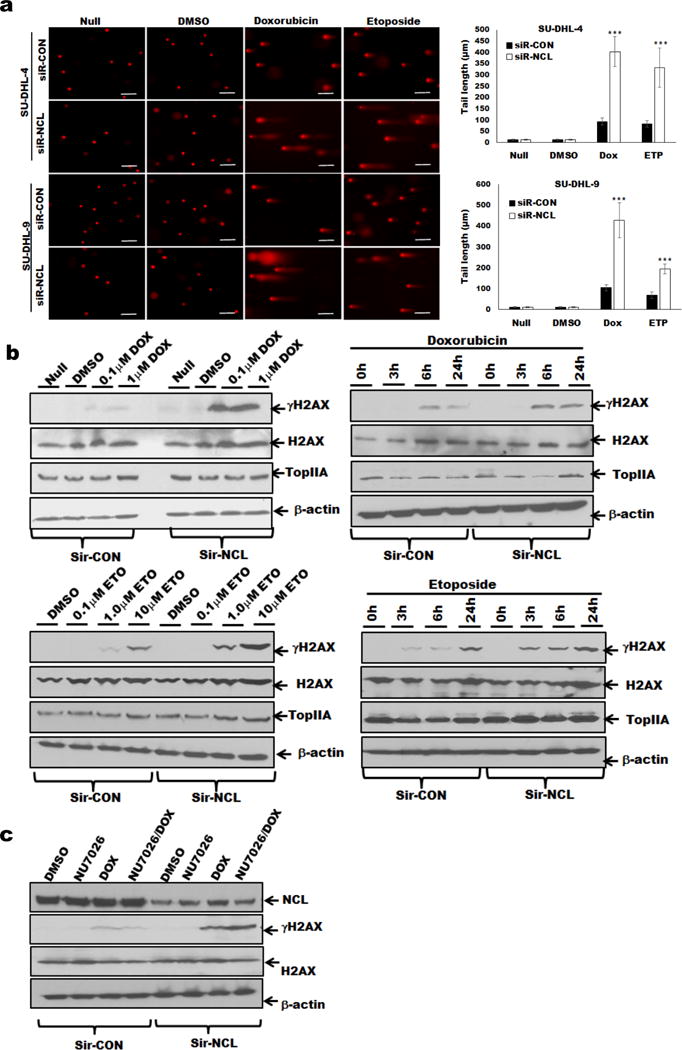
Nucleolin silencing and TopIIA targeting agent-induced DNA damage in DLBCL cells.
(a) SU-DHL-4, and-9 cells (siR-CON or siR-NCL) were treated with doxorubicin (0.1 μM), or etoposide (1.0μM), or 0.1% DMSO for 24 hours, and analyzed for DNA damage by comet assay. Comet tail length (μm) was determined from 100 cells using ImageJ software and mean values were plotted. (b) SU-DHL-9 cells were treated with doxorubicin or etoposide as indicated time and analyzed for γH2AX/H2AX expression. Nucleolin-silenced cells showed enhanced phosphorylation of H2AX in a dose and time-dependent manner. (c) SU-DHL-9 cells (siR-CON or siR-NCL) were treated with 0.1% DMSO, 1 μM NU7026 (DNA-dependent protein kinase inhibitor), 1μM doxorubicin, or both and analyzed for γH2AX/H2AX expression.
We further analyzed the effects of nucleolin in regulating TopIIA targeting agent-mediated DNA damage; we generated stable SU-DHL-9 and SU-DHL-4 nucleolin-knockdown cells by transducing them with a lentivirus expressing shRNA targeting the 3′UTR of nucleolin. We confirmed that shRNAs generated stable knockdowns, and were amenable to nucleolin reconstitution (Figures 4a and Supplementary Figure S3a). We transfected a full-length nucleolin (FLAG tagged) plasmid and selected with neomycin as described in “Methods” (Figure 4a). The sh-NCL-2-SU-DHL-9, and sh-NCL-2-SU-DHL-4 cells had reduced growth rate with G1 phase cell cycle arrest and increased expression of the cyclin-dependent kinase inhibitor p21 (Supplementary Figures S3b and S3c) compared to sh-CON cells. We confirmed that nucleolin knockdown cells showed restoration of cell proliferation by ectopic expression of full-length nucleolin cDNA (Figure 4b). We found that sh-NCL-2 cells were less clonogenic (Figure 4c) and grows smaller colonies in methylcellulose compared to sh-CON or sh-NCL-2+NCL cells (Supplementary Figure S3d). Thus, down-regulation of nucleolin expression inhibited growth of DLBCL cells. The results obtained with sh-NCL-2 cells (Fig. 4) were similar to those performed with siRNA knockdowns (Fig. 2). These acquired properties in sh-NCL-2 cells were reversed by exogenous nucleolin expression (sh-NCL-2+NCL cells) confirming that the effects were directly attributed to nucleolin (Figures 4d and 4e). There was more caspase3, PARP cleavage and reduced BID (22kDa) expression in doxorubicin-treated sh-NCL-2 cells than in sh-CON or sh-NCL-2+NCL cells (Figure 4f). Doxorubicin-induced phosphorylation of γH2AX was noticeably higher in sh-NCL-2 cells than in sh-CON or sh-NCL-2+NCL cells (Figures 4f and g). Thus, independent analyses confirm that nucleolin suppresses TopIIA targeting agent-induced apoptosis in DLBCL cells.
Figure 4.
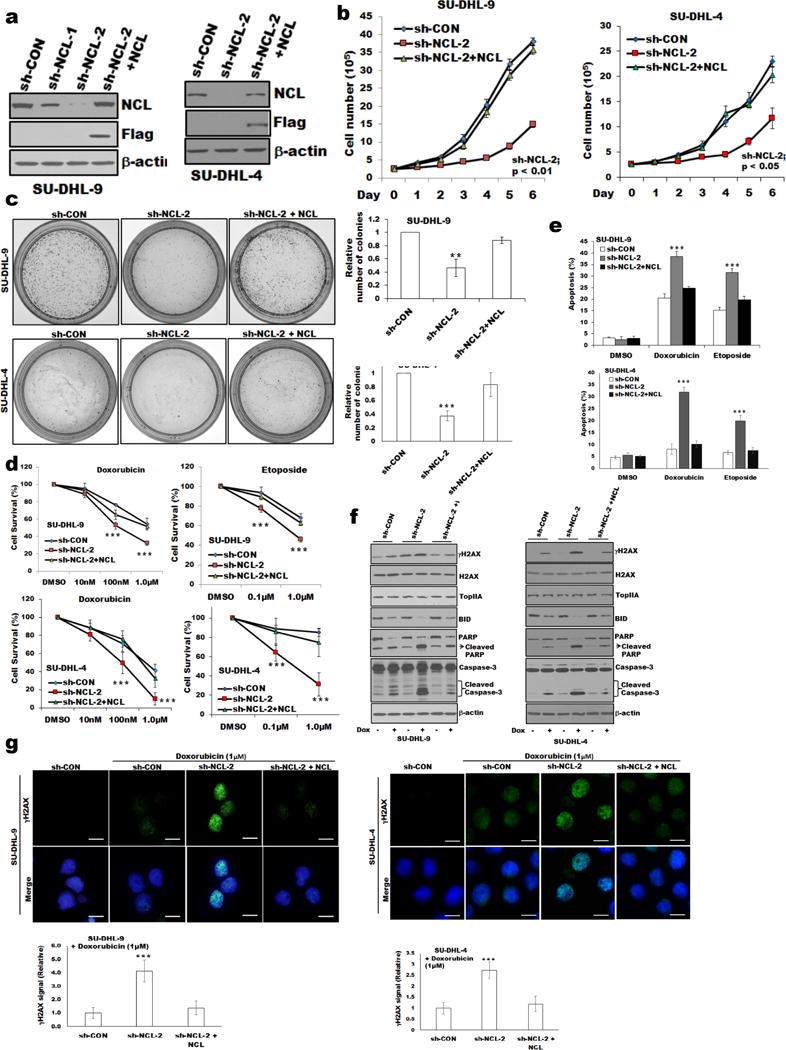
Proliferation and sensitization to doxorubicin-induced growth inhibition in nucleolin deregulated DLBCL cells.
(a) Stable knockdown of nucleolin expression in SU-DHL-4, and -9 cells using nucleolin-specific shRNAs (targeting 3′UTR). Reconstitution of nucleolin in nucleolin-knockdown cells (sh-NCL-2) was performed as described in “Methods”. Exogenous nucleolin expression in nucleolin knockdown cells was analyzed using anti-FLAG antibody. (b). Effect of nucleolin knockdown on cell proliferation. (c) Representative images of colony formation assay. The relative number of individual colonies in nucleolin-knockdown cells was significantly reduced compared to sh-control or sh-NCL-2+NCL cells, Bar=200μm. (d) MTT assay was performed as described in “Methods.” When treated with doxorubicin/etoposide, sh-NCL-2 cells showed reduced cell survival and increased percentage of apoptosis (e) compared to sh-control or sh-NCL-2+NCL cells. (f) Western blot of whole-cell lysates of SU-DHL-4, and -9 (sh-control or sh-NCL-2 or sh-NCL-2+NCL) cells treated with or without doxorubicin and analyzed for apoptotic signaling molecules, γH2AX/H2AX/TopIIA expression. (g) SU-DHL-4, and -9 cells (sh-control or sh-NCL-2 or sh-NCL-2+NCL) treated with doxorubicin were stained for γH2AX/DAPI and analyzed using fluorescence microscopy. Bar=20μm. The fluorescence intensity of γH2AX signal of 100 cells were calculated using ImageJ software and plotted. *p<0.05; **p<0.01; *** p<0.001
Nucleolin-TopIIA interaction regulates TopIIA targeting agent’s induced DNA damage
Because nucleolin is associates with nucleic acid and prevents apoptosis, we hypothesized that nucleolin might directly interact with TopIIA to regulate cell death. We first screened for an interaction between nucleolin and TopIIA. Co-immunoprecipitation analysis with either nucleolin or TopIIA antibodies followed by Western blotting in DLBCL cell lines (SU-DHL-4 and SU-DHL-9) confirmed the nucleolin-TopIIA association (Figure 5a).
Figure 5.
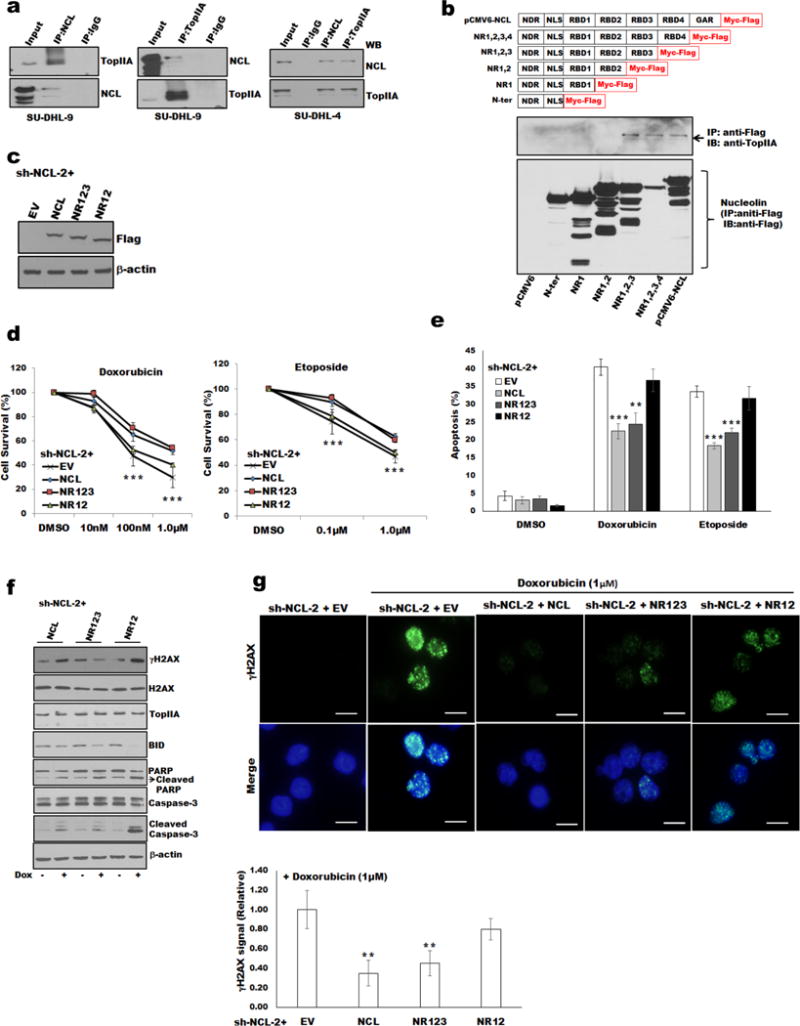
Nucleolin RBD3 domain and TopIIA complexes and doxorubicin induced DNA damage of DLBCL cells.
(a) Co-IP of nucleolin and TopIIA in whole-cell lysates of SU-DHL-4, and -9 cells. (b) Co-IP analysis and schematic of nucleolin and its functional domains: NDR, N-terminal domain region; NLS, nuclear localization signal; RBD1-4, RNA-binding domains 1–4; and GAR, glycine/arginine-rich C-terminal domain (upper panel). HEK293 cells were transfected with indicated nucleolin deletion mutants and lysed for IP/immunoblot analysis. (c) Western blot analysis for expression of transfected empty vector (EV), full-length nucleolin and deletion constructs (NR123 and NR12) in sh-NCL-2 SU-DHL-9 cells using anti-FLAG antibody. Expression of full-length nucleolin or deletion constructs NR123, but not NR12, rescued sh-NCL-2 SU-DHL-9 cells from doxorubicin/etoposide-induced cell survival (d) and apoptosis (e). (f) Expression of NR123 in sh-NCL-2 SU-DHL-9 cells protect cells from doxorubicin-induced apoptosis, and DNA damage determined by level of γH2AX by Western blot or staining/quantification with γH2AX antibody (g) Bar=20μm.**p<0.01; *** p<0.001
Having demonstrated the presence of TopIIA protein-nucleolin complexes, we embarked on mapping the domain of nucleolin required for binding to TopIIA by expressing nucleolin deletion mutants tagged with Myc-FLAG.12 This analysis revealed that only the mutants harboring RBD3 domain of nucleolin demonstrated the capacity to co-precipitate TopIIA (Figure 5b). To delineate the functional implications of the nucleolin-TopIIA interaction, we analyzed the effects of nucleolin mutants on doxorubicin-induced apoptosis and DNA damage in DLBCL cells deficient in endogenous nucleolin. The empty FLAG vector (EV) or FLAG tagged nucleolin constructs (full-length, NR123, NR12) were expressed in sh-NCL-2 cells. Western blot analysis using an anti-FLAG antibody confirmed the expression and levels of introduced nucleolin proteins (Figure 5c). Expression of nucleolin full-length or NR123, but not NR12, in sh-NCL-2 cells rescued the impaired survival and apoptosis when treated with doxorubicin or etoposide (Figures 5d and e). Moreover, phosphorylation of H2AX or cleavage/reduced of apoptosis markers (caspase 3, PARP, BID) after doxorubicin treatment was significantly increased in NR12-expressing cells but not in cells expressing nucleolin proteins capable of binding TopIIA (Figures 5f and g). These results support the concept of TopIIA-nucleolin complexes being a prerequisite for preventing TopIIA targeting agent-induced DNA damage and apoptosis.
Nucleolin regulates TopIIA activity
With these newly appointed properties of nucleolin on TopIIA, we characterized the effect of nucleolin on other TopIIA DNA repair functions. We measured the catalytic activity of TopIIA for decatenation of kinetoplast DNA (kDNA) with and without nucleolin. Catenated (interlinked) kDNA is the slow-moving band with minimal migration, as distinguished from decatenated (unlinked, monomer) kDNA, (Supplementary Figure S4a,II,III). We found less decatenated kDNA in extracts of nucleolin-silenced SU-DHL-9 cells than in control cells. Furthermore, we analyzed the effects of nucleolin on the DNA cleavage complexes formation. Treatment with etoposide induced cleavage of supercoiled pHOT-1 DNA and production of linear pHOT-1 in nuclear extracts (Supplementary Figure S4b). We found less supercoiled pHOT and more linear pHOT DNA in nucleolin-silenced nuclear extracts in the presence of etoposide than in siR-CON cells, suggesting that nucleolin silencing enhances TopIIA targeting agent-mediated DNA-cleavable complex formation.
To determine effects of nucleolin effect on TopIIA activity in DLBCL cells, we analyzed DNA-TopIIA complexes in cells exposed to etoposide. Cell extracts with chemically linked DNA-TopIIA were subjected to ultracentrifugation in a CsCl gradient and aliquots were blotted with anti-TopIIA antibody (Supplementary Figure S4c left panel). Treatment of cells with etoposide showed a dramatic shifted of TopIIA from a free protein fraction to a DNA bound fraction (lanes 9–10) in nucleolin-silenced cells. However, etoposide treatment failed to shift control cells, suggesting extensive binding of TopIIA to DNA after etoposide based treatment in nucleolin-silenced cells (Supplementary Figure S4c right panel). Phosphorylation of TopIIA affects TopIIA activity causing accumulation of TopIIA-DNA cleavable complex.31, 32 The results are consistent with accumulation of chemotherapy-induced, TopIIA-DNA complexes with enhanced phosphorylation of TopIIA (ser1469) levels in nucleolin-depleted cells (Supplementary Figure S4d).
Although TopIIA is highly expressed in cancers and its expression was not significantly correlated with clinical outcomes in DLBCL.33, 34 We performed immunohistochemistry (IHC) for TopIIA on the same 104 DLBCL patients reported in Figure 1. We observed no significant impact of TopIIA expression on OS (p=0.13) and PFS (p=0.17) of DLBCL patients (Supplementary Figure S4e). However, a significantly negative correlation of TopIIA expression with nucleolin levels was observed (Supplementary Figure S4f). Further analysis revealed that the DLBCL patients with nucleolinlow/TopIIAhigh had a significant (p<0.0001) better outcome (OS, PFS) compared to other groups of DLBCL patients (Supplementary Figure 4g). Our new data reported here indicates that TopIIA activity is modulated by nucleolin and the response of DLBCL cells to TopIIA targeting agents may depend on nucleolin TopIIA complexes.
Because nucleolin expresses oncogenic properties and is selectively expressed on the cancer cell surface, it has been a promising target for cancer therapies.35 We tested a nucleolin specific aptamer (AS1411) on DLBCL cell survival. We compared uptake of FL-AS1411, a fluorescently labeled version of the active aptamer, with that of FL-CRO, a fluorescently labeled control oligonucleotide with no anti-proliferative activity. FL-AS1411 uptake was more efficient by DLBCL cell lines (SU-DHL-4, SU-DHL-9) than control aptamer (Supplementary Figure S5a). Moreover, AS1411 treatment inhibited the cell survival and proliferation of DLBCL cells without affecting nucleolin expression (Supplementary Figures S5b–d). AS1411 treatment combined with doxorubicin/etoposide significantly inhibited cell survival of DLBCL cells (Figure 6a). DLBCL cells treated with nucleolin antagonist (Nucant) N6L (synthetic peptides, that targets surface nucleolin with high affinity and selectivity) induced cell death with some activity at sub-micro-molar doses (Figure 6b and Supplementary Figure S6). Thus, our results demonstrate that targeting nucleolin by several approaches improved the effects of chemotherapy.
Figure 6.
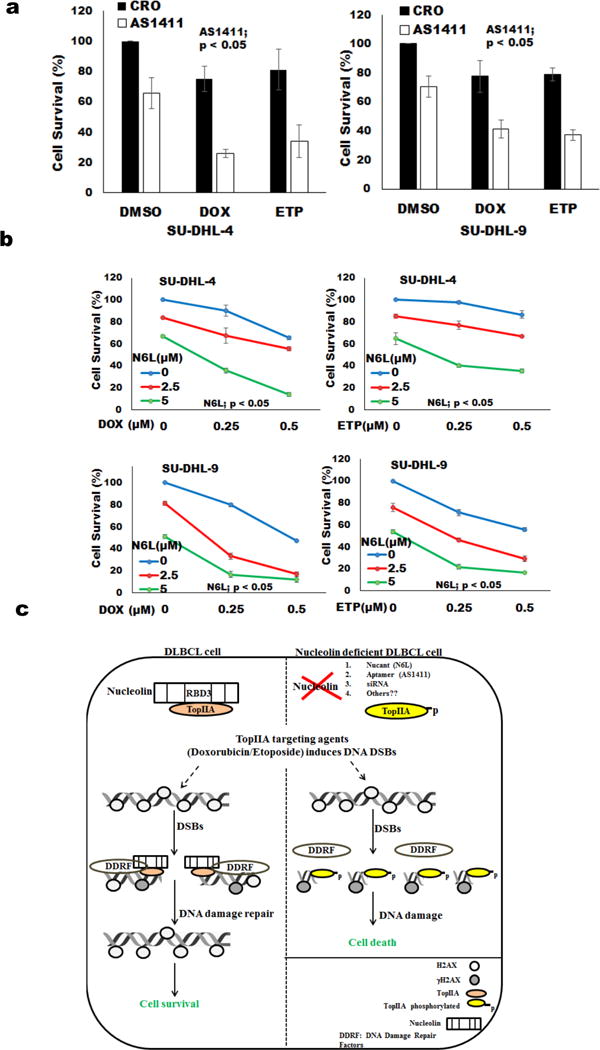
Nucleolin targeting agents in DLBCL.
(a) DLBCL cells were incubated with AS1411 (25μmol/l) or CRO for three days followed by adding doxorubicin or etoposide (0.1μM). Cell viability was analyzed using MTT assay after 24 hours of doxorubicin/etoposide treatment. (b) DLBCL cells were incubated with Nucant N6L along with either doxorubicin or etoposide. Cell viability was analyzed using MTT assay after 24 hours of treatment. (c) Model representing nucleolin function in DNA damage response: Nucleolin binds to TopIIA via its RBD3 and this nucleolin-TopIIA interaction resides at DSB and protects DLBCL cells from TopIIA targeting agent-mediated apoptosis. Silencing of nucleolin or blockage of nucleolin function (aptamer/nucant: AS1411/N6L or others) is associated with TopIIA targeting agent-induced accumulation of TopIIA-DNA cleavable complexes and enhances DLBCL cell apoptosis.
DISCUSSION
TopIIA has a key role of sensing and repairing damaged DNA3 and drugs that target TopIIA remain as critical components of therapy for lymphoma and leukemia. In this study, we found that TopIIA is regulated by nucleolin via nucleolin-TopIIA complex. This interaction promotes DNA repair and prevents apoptosis of DLBCL cells induced by TopIIA targeting agent (doxorubicin/etoposide) (Figure 6c). Silencing of nucleolin expression permits accumulation of of DNA damage and improves the killing effects of doxorubicin or etoposide (Figure 3 and 4). Moreover, inhibition of nucleolin activity by application of nucleolin specific aptamers (AS1411) or nucant (N6L) significantly decreased cell viability in the presence of doxorubicin (Figure 6 and Supplementary Figure 5 and 6). These findings are of clinical importance because, low versus high nucleolin levels in DLBCL predicted 90 month estimated survival of 70% versus 12% (P<0.0001) of patients treated with R-CHOP based therapy (Figure 1d and Supplementary Figure S1).
We found that depletion of nucleolin causes a robust accumulation of TopIIA-DNA complexes (Supplementary Figure S4c) and increased apoptosis of DLBCL cells after exposure to TopIIA targeting drugs (Figure 4e and f). The presence of nucleolin cleared TopIIA-DNA complexes from the cells suggesting that nucleolin was preventing DNA damage or facilitating DNA damage repair to overall promote DNA integrity and prevent apoptosis. These nucleolin functions were confirmed by reconstitution of nucleolin in nucleolin depleted cells. These nucleolin properties are not consider to occur secondary to nonspecific interactions of overexpressed protein, as the levels of introduced nucleolin and its derivative mutants were present at levels similar to those of endogenous nucleolin (Figure 4a and 5c).
Numerous interacting partners have shown to regulate the DNA repair function of TopIIA.3, 4, 31, 32, 36–39 In the present study, we observed that nucleolin silencing enhanced TopIIA targeting agent-induced DNA damage, as evidenced by DNA fragmentation accumulation in comet assay (Figure 3a) and by phosphorylation of H2AX26 and this effect was completely reversed by ectopic expression of nucleolin in nucleolin-silenced DLBCL cells (Figure 4g). Nucleolin is composed of an N-terminal domain rich in acidic residues, a central domain containing 4 RNA-binding motifs (RBD), and a C-terminal domain rich in arginine and glycine residues (RGG or GAR domain).40 RBD is known to bind the stem-loop structure of RNA and mediates processing of ribosomal RNA.40 We confirmed binding of nucleolin to TopIIA, and binding was restricted to RBD3 of nucleolin (Figure 5a and b) and binding is necessary for mediating effects on TopIIA functions. Our findings support the notion that nucleolin-TopIIA interaction regulates TopIIA targeting agent-mediated DNA damage and apoptosis of DLBCL cells, as the expression of a non-binding nucleolin deletion construct (NR12) failed to rescue TopIIA-mediated DNA damage and apoptosis in nucleolin-knockdown cells (Figure 5c–g).
DSBs generated by TopIIA targeting agents’ leads to immediate activation and recruitment of DNA repair components at DSB site. DNA damage response is mediated by protein sensors such as MRN (Mre11-Rad50-Nbs1) complexes, which trigger the activation of a signal transduction system including protein kinases ATM, ATR, Chk1, and p53.41,42 A previous report suggests that doxorubicin activates multiple ATM-dependent downstream targets required for DSBs repair.28 Conversely, inhibition of ATM blocks the etoposide-induced DNA damage response and apoptosis in human T-cells, suggesting that ATM regulated signaling is critical for DNA damage repair.43 In another cell model, nucleolin recruited to DSBs via interaction with RAD50 through its GAR domain and facilitate DNA DSB repair by recruiting repair factors at DSB sites.8 In our study, expression of RBD4-GAR deletion construct NR123 of nucleolin was able to rescue DNA damage/apoptosis defects in nucleolin-silenced DLBCL cells (Figure 5), Thus GAR is an optional domain for some DSB repairs. Our study is consistent to previous report where nucleolin-deficient cells exhibited increase γH2AX foci over control cells after ɤ-ray exposure. Accordingly, nucleolin appears as a participant in DNA repair regardless of source of damage.9
In this current study, nucleolin modulating of TopIIA function is novel. We found that nucleolin silencing decreased TopIIA decatenation activity and enhanced formation of TopIIA-DNA cleavable complexes in DLBCL cells in the presence of etoposide (Supplementary Figure S4a–c) without affecting the TopIIA levels. This suggests that nucleolin as an interacting partner of TopIIA might be able to regulate its enzymatic activity by resolving DSB. This postulated regulation is consistent with previous reports indicating TopIIA interacting partners profoundly affect TopIIA enzyme activity.3 For example, ATM and casein kinase I (CKI) enhance etoposide-stabilized TopIIA-DNA-cleavable complex formation,31,32 and casein Kinase II (CKII) enhances decatenation activity of TopIIA.36, 37 TopIIA itself serves as a substrate of many kinases, which phosphorylate and influence its enzymatic activity.31, 32 In this study, we found that nucleolin silencing enhanced the phosphorylation of TopIIA at serine residue 1469, which is one of the mechanisms to alter TopIIA activity and enhance TopIIA-DNA cleavable complex formation (Supplementary Figure S4c and d). We speculate that nucleolin silencing might reduce the competition for binding to TopIIA, rendering TopIIA available to other kinases and their regulation.
High levels of TopIIA are associated in DLBCL patients who have improved clinical response to anthracycline-based chemotherapy.33, 34 This clinical benefit stands in agreement with DNA repair capabilities provided by TopIIA. We observed no significant impact of TopIIA expression on OS and PFS of DLBCL patients (Supplementary Figure S4e). Nucleolin is an important target that exerts prosurvival effect expected to transcend those mediated in partnership with TopIIA. Nucleolin dictates the expression of many genes such as Bcl-xL, AKT, and IL-2 that regulate apoptosis and support the transformed cell phenotype.14,44,45 In chronic lymphocytic leukemia, a 26-fold elevated level of nucleolin support a 11-fold bcl-2 protein levels by protecting bcl-2 mRNA.14 Thus, nucleolin levels may serve as a predictive biomarker of anthracycline/etoposide sensitivity and a marker for survival with other apoptosis stimuli.
Nucleolin is highly expressed on surface of several types of cancer rendering it an attractive target for cancer therapies.12, 13, 35, 46 Earlier we demonstrated that primary non-Hodgkin lymphoma including DLBCL expressed nucleolin on the cell surface whereas normal B cells lacked expression.12 Nucleolin on cancer cell surfaces has gain recognition for having diverse functions such as shuttling and chaperone function for ligands, cytokines, acting as receptor for viruses and regulating receptor functions.10–14, 47–51 We have shown that silencing of nucleolin expression preferentially depletes the cell surface nucleolin and speculate that lack of surface nucleolin is sufficient to alter transmembrane signaling.12 A number of molecules such as aptamers (AS1411) or peptides/immuno-agents (HB19, N6L, F3, 4LB5) have been known to inhibit nucleolin function and rely on access to cell surface nucleolin.52,53,54 AS1411 is the first DNA aptamer that has reach phase I and II clinical trials for potential treatment of cancer including acute myelogenous leukemia (AML).55 Moreover, AS1411 kill AML cells from patients at 5μM, without affecting normal B cell.56, 57 AS1411 conjugated to cytotoxic agents, showed selective delivery of these agents to tumor cells. For example, gemcitabine (GEM)-loaded AS1411 aptamer surface-decorated nanopolymersome has been used in non-small cell lung cancer.58 In mice xenografts, AS1411-doxorubicin conjugation has shown a specific uptake/release of drug in hepatocellular carcinoma and reduced tumor formation.59 We found combined nucleolin-specific AS1411 and doxorubicin or etoposide treatment significantly inhibited the survival and proliferation of DLBCL cells (Figure 6a and Supplementary Figure 5). Similar results were obtained when DLBCL cells were treated with nucleolin specific nucant N6L (Figure 6b and Supplementary Figure 6). N6L treatment has strongly inhibited breast cancer growth by inducing apoptosis60 and is currently aim for phase II clinical trials (IPP-204106).
In conclusion, nucleolin binds to TopIIA and this interaction blocks the effects of TopIIA targeting agent-induced DNA damage and apoptosis of DLBCL. The efficacy of TopIIA targeting agents currently in clinical use might be enhanced by blocking nucleolin in DLBCL cells. We anticipate targeting nucleolin by several approaches would improve the effects of chemotherapy for DLBCL.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute grant (CA 1206173), National Institute of Diabetes and Digestive and Kidney Diseases grant (DK091490), the Richard Spencer Lewis Memorial Foundation, and patients’ families. The MD Anderson Flow Cytometry and Cellular Imaging Facility are supported by the NIH/NCI under award number P30CAQ16672.
Footnotes
CONFLICT OF INTEREST: The authors declare no conflict of interest
Contribution: N.J., H.F.Z, L.S. & F.S. designed the research studies, analyzed the data, and wrote the manuscript. N.J., H.F.Z., B.G., T.K., Q.Y. R.M., R.K.S, F.K.B., X.W., Z.B., J.W., X.Y.X.M., K.H.Y. performed experiments, analyzed the data and contributed clinical resources under approved IRB. Q.Y. and K.H.Y. analyzed the tissue staining, quantified protein expression and identified correlations with patient clinical outcome. L.S. and F.S. contributed to the research design, writing and finalized the manuscript.
References
- 1.Friedberg JW, Fisher RI. Diffuse large B-cell lymphoma. Hematol Oncol Clin North Am. 2008 Oct;22(5):941–952, ix. doi: 10.1016/j.hoc.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics, 2012. CA: a cancer journal for clinicians. 2012 Jul-Aug;62(4):220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 3.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009 May;9(5):327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chikamori K, Grozav AG, Kozuki T, Grabowski D, Ganapathi R, Ganapathi MK. DNA topoisomerase II enzymes as molecular targets for cancer chemotherapy. Curr Cancer Drug Targets. 2010 Nov;10(7):758–771. doi: 10.2174/156800910793605785. [DOI] [PubMed] [Google Scholar]
- 5.Arriola E, Rodriguez-Pinilla SM, Lambros MB, Jones RL, James M, Savage K, et al. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast Cancer Res Treat. 2007 Dec;106(2):181–189. doi: 10.1007/s10549-006-9492-5. [DOI] [PubMed] [Google Scholar]
- 6.Press MF, Sauter G, Buyse M, Bernstein L, Guzman R, Santiago A, et al. Alteration of topoisomerase II-alpha gene in human breast cancer: association with responsiveness to anthracycline-based chemotherapy. J Clin Oncol. 2011 Mar 1;29(7):859–867. doi: 10.1200/JCO.2009.27.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scandinavian Breast Group T. Tanner M, Isola J, Wiklund T, Erikstein B, Kellokumpu-Lehtinen P, et al. Topoisomerase IIalpha gene amplification predicts favorable treatment response to tailored and dose-escalated anthracycline-based adjuvant chemotherapy in HER-2/neu-amplified breast cancer: Scandinavian Breast Group Trial 9401. J Clin Oncol. 2006 Jun 1;24(16):2428–2436. doi: 10.1200/JCO.2005.02.9264. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB. Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proc Natl Acad Sci U S A. 2013 Oct 15;110(42):16874–16879. doi: 10.1073/pnas.1306160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi J, Fujimoto H, Sato J, Hayashi I, Burma S, Matsuura S, et al. Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway. PLoS One. 2012;7(11):e49245. doi: 10.1371/journal.pone.0049245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogal V, Sugahara KN, Ruoslahti E, Christian S. Cell surface nucleolin antagonist causes endothelial cell apoptosis and normalization of tumor vasculature. Angiogenesis. 2009;12(1):91–100. doi: 10.1007/s10456-009-9137-5. [DOI] [PubMed] [Google Scholar]
- 11.Pichiorri F, Palmieri D, De Luca L, Consiglio J, You J, Rocci A, et al. In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. J Exp Med. 2013 May 6;210(5):951–968. doi: 10.1084/jem.20120950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wise JF, Berkova Z, Mathur R, Zhu H, Braun FK, Tao RH, et al. Nucleolin inhibits Fas ligand binding and suppresses Fas-mediated apoptosis in vivo via a surface nucleolin-Fas complex. Blood. 2013 Jun 6;121(23):4729–4739. doi: 10.1182/blood-2012-12-471094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu W, Zhou F, Zhang Q, Sun X, Shi X, Liang Y, et al. Overexpression of nucleolin and different expression sites both related to the prognosis of gastric cancer. APMIS. 2013 Oct;121(10):919–925. doi: 10.1111/apm.12131. [DOI] [PubMed] [Google Scholar]
- 14.Otake Y, Soundararajan S, Sengupta TK, Kio EA, Smith JC, Pineda-Roman M, et al. Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood. 2007 Apr 1;109(7):3069–3075. doi: 10.1182/blood-2006-08-043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bharti AK, Olson MO, Kufe DW, Rubin EH. Identification of a nucleolin binding site in human topoisomerase I. J Biol Chem. 1996 Jan 26;271(4):1993–1997. doi: 10.1074/jbc.271.4.1993. [DOI] [PubMed] [Google Scholar]
- 16.Yang C, Kim MS, Chakravarty D, Indig FE, Carrier F. Nucleolin Binds to the Proliferating Cell Nuclear Antigen and Inhibits Nucleotide Excision Repair. Molecular and cellular pharmacology. 2009;1(3):130–137. doi: 10.4255/mcpharmacol.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain N, Thanabalu T. Molecular difference between WASP and N-WASP critical for chemotaxis of T-cells towards SDF-1alpha. Scientific reports. 2015;5:15031. doi: 10.1038/srep15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathur R, Chandna S, P NK, B SD. Peptidyl prolyl isomerase, Pin1 is a potential target for enhancing the therapeutic efficacy of etoposide. Curr Cancer Drug Targets. 2011 Mar;11(3):380–392. doi: 10.2174/156800911794519761. [DOI] [PubMed] [Google Scholar]
- 19.Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Li Y, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012 Sep;26(9):2103–2113. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visco C, Tzankov A, Xu-Monette ZY, Miranda RN, Tai YC, Li Y, et al. Patients with diffuse large B-cell lymphoma of germinal center origin with BCL2 translocations have poor outcome, irrespective of MYC status: a report from an International DLBCL rituximab-CHOP Consortium Program Study. Haematologica. 2013 Feb;98(2):255–263. doi: 10.3324/haematol.2012.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu-Monette ZY, Wu L, Visco C, Tai YC, Tzankov A, Liu WM, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2012 Nov 08;120(19):3986–3996. doi: 10.1182/blood-2012-05-433334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006 Jun 08;354(23):2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 23.Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008 Nov 27;359(22):2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathur R, Sehgal L, Havranek O, Kohrer S, Khashab T, Jain N, et al. Inhibition of demethylase KDM6B sensitizes diffuse large B-cell lymphoma to chemotherapeutic drugs. Haematologica. 2017 Feb;102(2):373–380. doi: 10.3324/haematol.2016.144964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelmohsen K, Gorospe M. RNA-binding protein nucleolin in disease. RNA Biol. 2012 Jun;9(6):799–808. doi: 10.4161/rna.19718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010 Apr;24(4):679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 27.Karpinich NO, Tafani M, Schneider T, Russo MA, Farber JL. The course of etoposide-induced apoptosis in Jurkat cells lacking p53 and Bax. Journal of cellular physiology. 2006 Jul;208(1):55–63. doi: 10.1002/jcp.20638. [DOI] [PubMed] [Google Scholar]
- 28.Kurz EU, Douglas P, Lees-Miller SP. Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. The Journal of biological chemistry. 2004 Dec 17;279(51):53272–53281. doi: 10.1074/jbc.M406879200. [DOI] [PubMed] [Google Scholar]
- 29.Schonn I, Hennesen J, Dartsch DC. Ku70 and Rad51 vary in their importance for the repair of doxorubicin- versus etoposide-induced DNA damage. Apoptosis : an international journal on programmed cell death. 2011 Apr;16(4):359–369. doi: 10.1007/s10495-010-0564-y. [DOI] [PubMed] [Google Scholar]
- 30.Willmore E, de Caux S, Sunter NJ, Tilby MJ, Jackson GH, Austin CA, et al. A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood. 2004 Jun 15;103(12):4659–4665. doi: 10.1182/blood-2003-07-2527. [DOI] [PubMed] [Google Scholar]
- 31.Grozav AG, Chikamori K, Kozuki T, Grabowski DR, Bukowski RM, Willard B, et al. Casein kinase I delta/epsilon phosphorylates topoisomerase IIalpha at serine-1106 and modulates DNA cleavage activity. Nucleic Acids Res. 2009 Feb;37(2):382–392. doi: 10.1093/nar/gkn934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritke MK, Murray NR, Allan WP, Fields AP, Yalowich JC. Hypophosphorylation of topoisomerase II in etoposide (VP-16)-resistant human leukemia K562 cells associated with reduced levels of beta II protein kinase C. Mol Pharmacol. 1995 Nov;48(5):798–805. [PubMed] [Google Scholar]
- 33.Pentheroudakis G, Goussia A, Voulgaris E, Nikolaidis K, Ioannidou E, Papoudou-Bai A, et al. High levels of topoisomerase IIalpha protein expression in diffuse large B-cell lymphoma are associated with high proliferation, germinal center immunophenotype, and response to treatment. Leuk Lymphoma. 2010 Jul;51(7):1260–1268. doi: 10.3109/10428194.2010.483749. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Wang J, Zhang H, Liu D, Li Y, Xu Y, et al. Topo IIalpha gene alterations correlated with survival in patients with diffuse large B-cell lymphoma. Eur J Clin Invest. 2012 Mar;42(3):310–320. doi: 10.1111/j.1365-2362.2011.02585.x. [DOI] [PubMed] [Google Scholar]
- 35.Koutsioumpa M, Papadimitriou E. Cell surface nucleolin as a target for anti-cancer therapies. Recent Pat Anticancer Drug Discov. 2014 May;9(2):137–152. doi: 10.2174/1574892808666131119095953. [DOI] [PubMed] [Google Scholar]
- 36.Cardenas ME, Walter R, Hanna D, Gasser SM. Casein kinase II copurifies with yeast DNA topoisomerase II and re-activates the dephosphorylated enzyme. J Cell Sci. 1993 Feb;104(Pt 2):533–543. doi: 10.1242/jcs.104.2.533. [DOI] [PubMed] [Google Scholar]
- 37.Alghisi GC, Roberts E, Cardenas ME, Gasser SM. The regulation of DNA topoisomerase II by casein kinase II. Cell Mol Biol Res. 1994;40(5–6):563–571. [PubMed] [Google Scholar]
- 38.Lou Z, Minter-Dykhouse K, Chen J. BRCA1 participates in DNA decatenation. Nat Struct Mol Biol. 2005 Jul;12(7):589–593. doi: 10.1038/nsmb953. [DOI] [PubMed] [Google Scholar]
- 39.Kurz EU, Leader KB, Kroll DJ, Clark M, Gieseler F. Modulation of human DNA topoisomerase IIalpha function by interaction with 14-3-3epsilon. J Biol Chem. 2000 May 5;275(18):13948–13954. doi: 10.1074/jbc.275.18.13948. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava M, Pollard HB. Molecular dissection of nucleolin’s role in growth and cell proliferation: new insights. FASEB J. 1999 Nov;13(14):1911–1922. [PubMed] [Google Scholar]
- 41.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009 Oct 22;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nature reviews Cancer. 2008 Jul;8(7):512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 43.Korwek Z, Sewastianik T, Bielak-Zmijewska A, Mosieniak G, Alster O, Moreno-Villanueva M, et al. Inhibition of ATM blocks the etoposide-induced DNA damage response and apoptosis of resting human T cells. DNA repair. 2012 Nov 1;11(11):864–873. doi: 10.1016/j.dnarep.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Chen CY, Gherzi R, Andersen JS, Gaietta G, Jurchott K, Royer HD, et al. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000 May 15;14(10):1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 45.Gao G, Dou QP. G(1) phase-dependent expression of bcl-2 mRNA and protein correlates with chemoresistance of human cancer cells. Molecular pharmacology. 2000 Nov;58(5):1001–1010. doi: 10.1124/mol.58.5.1001. [DOI] [PubMed] [Google Scholar]
- 46.Berger CM, Gaume X, Bouvet P. The roles of nucleolin subcellular localization in cancer. Biochimie. 2015 Jun;113:78–85. doi: 10.1016/j.biochi.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 47.Hovanessian AG. Midkine, a cytokine that inhibits HIV infection by binding to the cell surface expressed nucleolin. Cell Res. 2006 Feb;16(2):174–181. doi: 10.1038/sj.cr.7310024. [DOI] [PubMed] [Google Scholar]
- 48.Said EA, Courty J, Svab J, Delbe J, Krust B, Hovanessian AG. Pleiotrophin inhibits HIV infection by binding the cell surface-expressed nucleolin. FEBS J. 2005 Sep;272(18):4646–4659. doi: 10.1111/j.1742-4658.2005.04870.x. [DOI] [PubMed] [Google Scholar]
- 49.Chen YL, Liu CD, Cheng CP, Zhao B, Hsu HJ, Shen CL, et al. Nucleolin is important for Epstein-Barr virus nuclear antigen 1-mediated episome binding, maintenance, and transcription. Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):243–248. doi: 10.1073/pnas.1321800111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balinsky CA, Schmeisser H, Ganesan S, Singh K, Pierson TC, Zoon KC. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J Virol. 2013 Dec;87(24):13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med. 2011 Sep;17(9):1132–1135. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
- 52.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol. 2009 Jun;86(3):151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilles ME, Maione F, Cossutta M, Carpentier G, Caruana L, Di Maria S, et al. Nucleolin Targeting Impairs the Progression of Pancreatic Cancer and Promotes the Normalization of Tumor Vasculature. Cancer Res. 2016 Dec 15;76(24):7181–7193. doi: 10.1158/0008-5472.CAN-16-0300. [DOI] [PubMed] [Google Scholar]
- 54.Krust B, El Khoury D, Nondier I, Soundaramourty C, Hovanessian AG. Targeting surface nucleolin with multivalent HB-19 and related Nucant pseudopeptides results in distinct inhibitory mechanisms depending on the malignant tumor cell type. BMC Cancer. 2011 Aug 03;11:333. doi: 10.1186/1471-2407-11-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mongelard F, Bouvet P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr Opin Mol Ther. 2010 Feb;12(1):107–114. [PubMed] [Google Scholar]
- 56.Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther. 2006 Dec;5(12):2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 57.Damian Laber BT, Goetz Kloecker, Gary Acton, Donald Miller. Pharmacokinetics of the anti-nucleolin aptamer AS1411 in a phase I study. American Association for Cancer Research; San Francisco, CA: 2007. [Google Scholar]
- 58.Alibolandi M, Ramezani M, Abnous K, Hadizadeh F. AS1411 Aptamer-Decorated Biodegradable Polyethylene Glycol-Poly(lactic-co-glycolic acid) Nanopolymersomes for the Targeted Delivery of Gemcitabine to Non-Small Cell Lung Cancer In Vitro. Journal of pharmaceutical sciences. 2016 May;105(5):1741–1750. doi: 10.1016/j.xphs.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 59.Trinh TL, Zhu G, Xiao X, Puszyk W, Sefah K, Wu Q, et al. A Synthetic Aptamer-Drug Adduct for Targeted Liver Cancer Therapy. PLoS One. 2015;10(11):e0136673. doi: 10.1371/journal.pone.0136673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Destouches D, Page N, Hamma-Kourbali Y, Machi V, Chaloin O, Frechault S, et al. A simple approach to cancer therapy afforded by multivalent pseudopeptides that target cell-surface nucleoproteins. Cancer Res. 2011 May 01;71(9):3296–3305. doi: 10.1158/0008-5472.CAN-10-3459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


