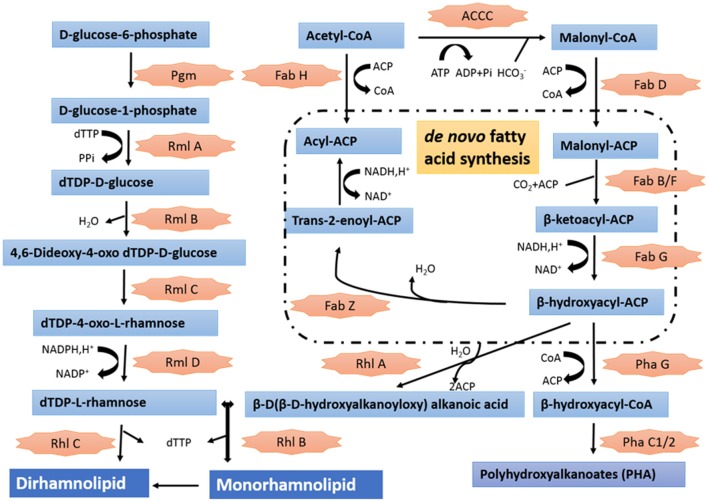
Figure 4.
Putative metabolic pathway involved in the synthesis of rhamnolipid in genome of P. rhisophila S211. Pgm, Phosphoglucomutase, EC 5.4.2.2; RmlA, Glucose-1-phosphate thymidylyltransferase, EC 2.7.7.24; RmlB, dTDP-glucose 4,6-dehydratase, EC 4.2.1.46; RmlC, dTDP-4-dehydrorhamnose 3,5-epimerase, EC 5.1.3.13; RmlD, dTDP-4-dehydrorhamnose reductase, EC 1.1.1.133; RhlB and RhlC, Glycosyltransferase, EC 2.4.1.; PhaG, (R)-3-hydroxydecanoyl-ACP:CoA transacylase PhaG (3-hydroxyacyl-CoA-acyl carrier protein transferase), EC 2.4.1.-; PhaC1/2: Polyhydroxyalkanoic acid synthases, EC 2.3.1-; ACCC, Acetyl-coenzyme A carboxyl transferase alpha chain, EC 6.4.1.2; FabD, Malonyl CoA-acyl carrier protein transacylase, EC 2.3.1.39; Fab H, 3-oxoacyl-[acyl-carrier-protein] synthase, KASIII, EC 2.3.1.180; FabB/F, 3-oxoacyl-[ACP] synthase, EC 2.3.1.41; FabG, 3-oxoacyl-[acyl-carrier protein] reductase, EC 1.1.1.100.