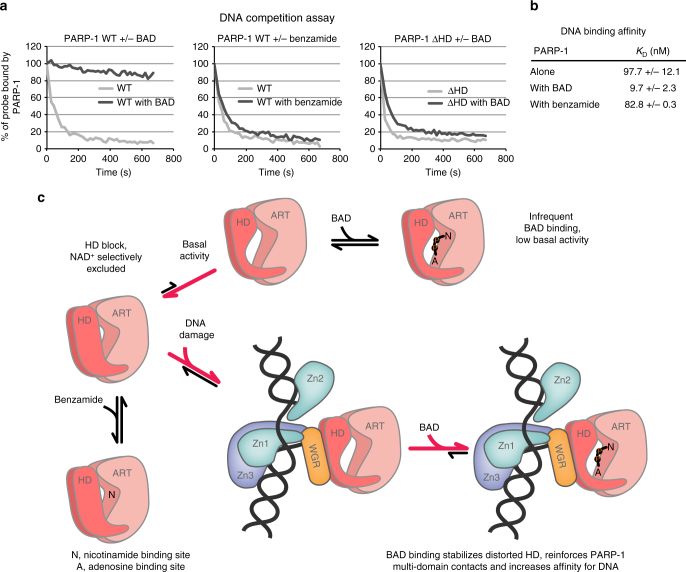
Fig. 6.
Allosteric coupling of PARP-1 catalytic active site and DNA-binding domains and model for the mechanism of HD regulation. a The fluorescence polarization DNA competition assay was performed by incubating 40 nM of PARP-1 WT or PARP-1 ΔHD with 20 nM of fluorescently labeled DNA (dumbbell DNA containing a central nick and an internal fluorescent FAM group) in the absence or presence of BAD or benzamide at 500 μM. A competitor unlabeled DNA was added and FP was measured over time. A representative experiment is shown. b PARP-1 affinity for DNA in the presence or absence of the indicated compounds, as measured by fluorescence polarization (see also Supplementary Fig. 9). c In its native state, the HD primarily exists in a folded conformation that selectively restricts access to the NAD+-binding site, which can be subdivided into the nicotinamide binding site (N) and the adenosine binding site (A). Small molecules such as benzamide can access the NAD+-binding site in the closed HD state, but BAD and NAD+ binding is completely blocked in the folded state of the HD. The inherent dynamics of the HD sample the open conformation infrequently, leading to the basal level of PARP-1 catalytic output. The DNA-damage-dependent regulatory domains of PARP-1 form an interface with the HD that promotes an open, unfolded HD conformation that allows NAD+ to have access to the active site. BAD binding to the catalytic active site shifts the HD equilibrium distribution to the unfolded state, thereby promoting the multi-domain contacts of PARP-1 with damage DNA, and increasing PARP-1 affinity for DNA (see panels a and b)