Figure 2.
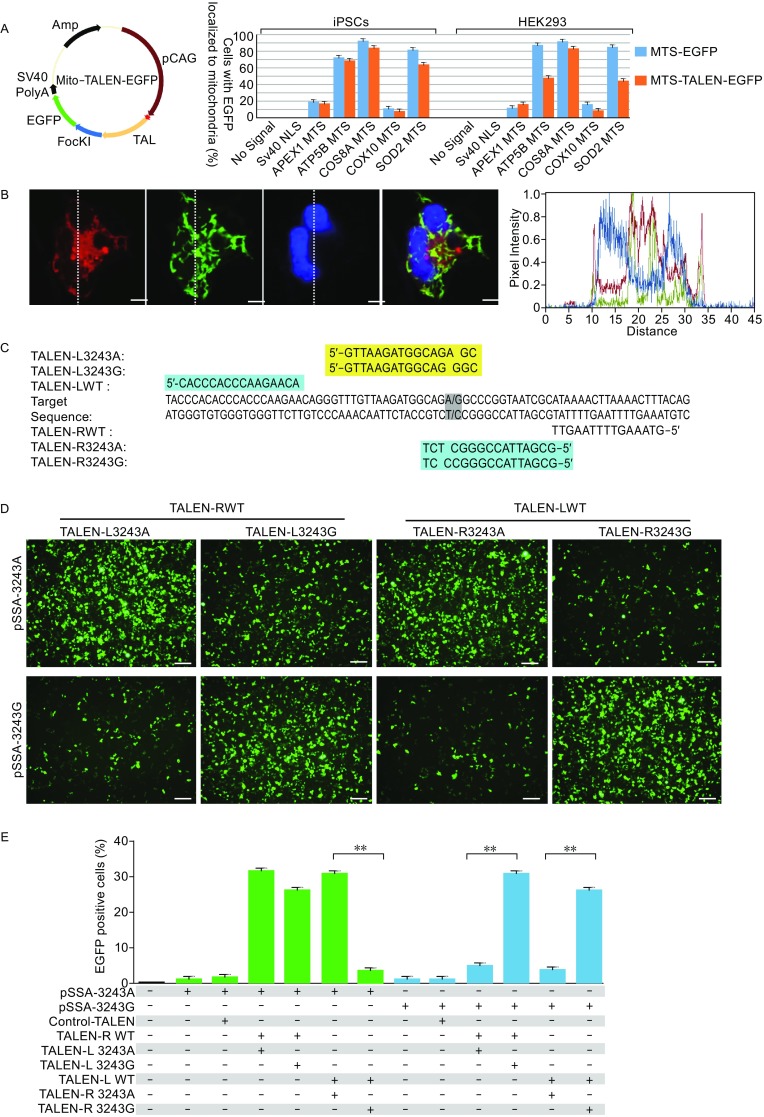
Engineering of mitoTALENs that specifically target 3243G mutant mtDNA. (A) The specificity of mitochondrial localization of EGFP and the TALEN-EGFP fusion proteins mediated by MTS derived from different nuclear genes. Left, schematic drawing of the engineered TALEN-EGFP polypeptide monomer containing an MTS in the N-terminus (MLS). The specificity and efficiency of MTS targeting to the mitochondria were analyzed based on the percentage of the EGFP and MitoTracker co-localizing cells in all of the transfected iPSCs and HEK293 cells, as shown in Fig. S1B and S1C. (B) Mitochondrial localization of mitoTALEN monomers mediated by COX8A-MTS in iPSCs 24 h after transfection. Mitochondria were visualized by MitoTracker Red. Scale bar = 10 µmol/L. Co-localization was visualized by the overlapping peaks of the relative fluorescence intensity (y-axis) on lines that passed through areas with marker signal. The position of the lines is indicated on the images, with lines running from top to bottom. (C) TALENs designed for targeting the mtDNA 3243 locus. (D and E) A single-strand annealing (SSA) assay to determine the specific targeting of the 3243G mtDNA mutation by TALENs. The mutated EGFP coding sequence was divided into two segments, which were separated by a stop codon and targeting sequence. Both segments contained an identical homology region. Once double-strand breaks were introduced into the target site by TALENs, the mutated EGFP coding sequence was repaired by annealing the two homologous sequences. Expression of EGFP was detected with a confocal microscope using appropriate filters after 48 h (D), (scale bars 50 μm), and the proportion of the EGFP-positive cells was measured by flow cytometry (E) (n = 3, error bars represent ±SEM; **P < 0.05)
