Figure 2.
Changes in H3K9ac and H3K9me3 at Bivalently Marked Promoters
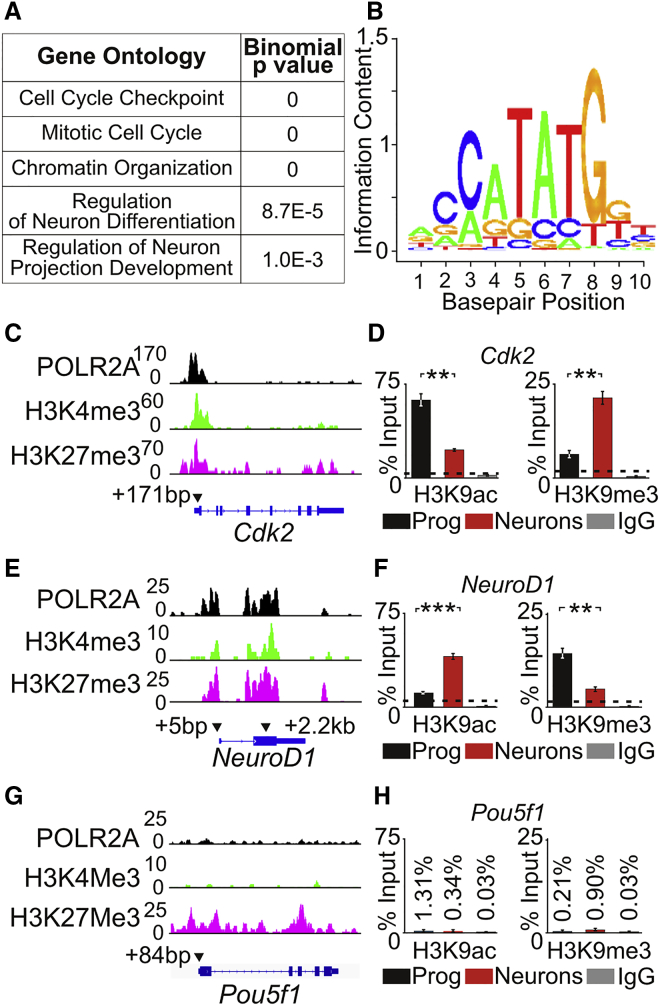
(A) Biological processes of genes with bivalent promoters were determined by gene ontology analysis. Pertinent biological process and associated binomial p values are listed.
(B) A consensus logo plot for NEUROG1 binding site containing an E-box (CANNTG).
(C) ChIP-seq peaks for POLR2A, H3K4me3, and H3K27me27 for Cdk2. Arrowheads indicate the approximate location of primers used for ChIP-qPCR. The number indicates the relative base-pair distance from the TSS.
(D) Enrichment of H3K9ac (active) or H3K9me3 (repressive) marks at the Cdk2 promoter region by ChIP-qPCR in proliferating iMOP (n = 3) and iMOP-derived neurons (n = 3). Background levels were determined by performing ChIP-qPCR with non-specific rabbit IgG control antibody.
(E) ChIP-seq peaks for POLR2A, H3K4me3, and H3K27me27 at NeuroD1. Arrowheads indicate the approximate location of primers relative to the TSS used for ChIP-qPCR.
(F) ChIP-qPCR of H3K9ac and H3K9me3 at the NeuroD1 promoter in proliferating progenitor (n = 3) and iMOP-derived neurons (n = 3).
(G) ChIP-seq peaks for POLR2A, H3K4me3, and H3K27me27 at Pou5f1.
(H) ChIP-qPCR of H3K9ac and H3K9me3 at the Pou5f1 promoter in proliferating progenitors (n = 3) and iMOP-derived neurons (n = 3).
Error bars denote ±SEM. ∗∗p < 0.01, ∗∗∗p < 0.001.