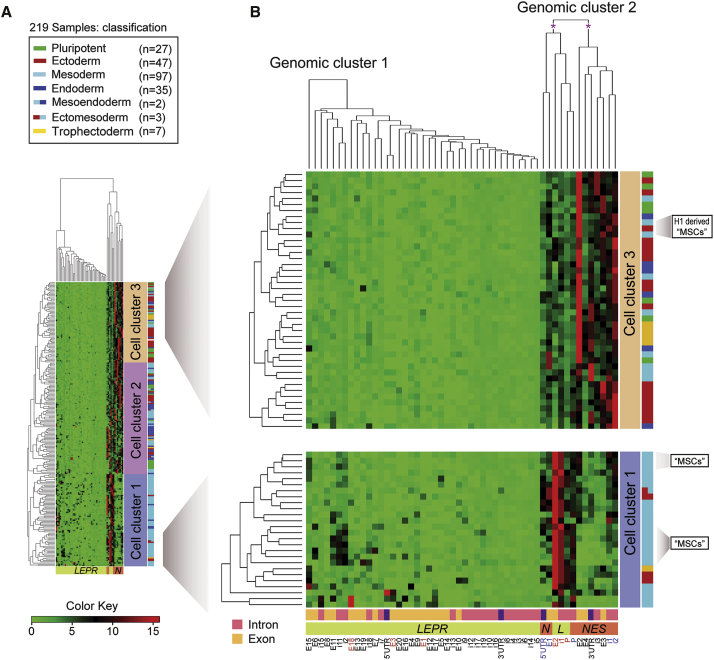
Figure 4.
Representative Analysis of the Epigenetic Marker H3K4me1 at the NES and LEPR Loci
(A) Clustering analysis of deposited H3K4me1 ChiP-seq data (www.genboree.org) in 219 samples that comprise cell types from three germ layers and trophectoderm (Table S2, Figure S1). H3K4me1 data for 219 human samples (GEO accession number: GGSM621418) were imported into the Genboree Workbench from Release 9 of the Human Epigenome Atlas (www.genboree.org). Human genome assembly GRCh37/hg19 (February 2009) was used for this analysis. The normalized values for NES and LEPR were exported for cluster analysis and visualization in R (www.cran.r-project.org) using the heatmap.2 function.
(B) The enlarged views of the regions of interest are presented on the right panel. Asterisks indicate the views of truncated dendrograms. Detailed information for these dendrograms is available from Figure S1. Of note, the genomic localization of exon 20 of the LEPR gene is currently not available from the Human Epigenome Atlas (www.genboree.org). Hence, the epigenomic data of exon 20 should be interpreted with caution.
H1, human embryonic stem cell line H1 (WA01); H3K4me1, monomethylated histone H3 lysine 4; I, intron; LEPR or L, the leptin receptor gene; LEPROT or Leprot, leptin receptor overlapping transcript; “MSC,” “mesenchymal stem cells”; NES or N, the gene coding for nestin; UTR, untranslated region.