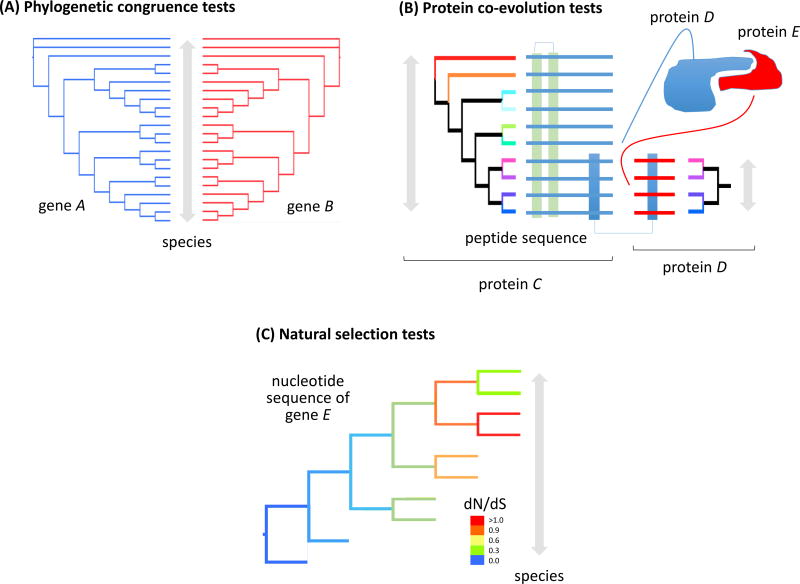
Figure 5. Statistical tests for phylogenetic congruence, protein co-evolution, and natural selection.
(A) Phylogenetic congruence tests measure and visualize the extent to which tree topologies differ. These tests can be used to provide evidence for differing patterns of evolution among genes, as well as gene duplication, horizontal gene transfer, or other nonvertical inheritance. Phylogenetic congruence tests may be classified as character congruence tests, of which the incongruence length difference test is most well-known, and topological congruence tests, including the tanglegram shown here (the gene A and B topologies are congruent). (B) Tests for protein co-evolution measure the coordinated changes that occur in pairs of proteins or protein residues, typically to maintain or refine catalytic or ligand-binding interactions (between protein C and D; modified from de Juan et al. 2013). (C) dN/dS is an indicator of natural selection acting on a genetic locus. It is the ratio of mutations that change amino acids (nonsynonymous mutations) to those that do not (synonymous mutations). If selection is absent, and mutations are caused by random genetic drift, dN/dS = 1. dN/dS > 1 suggests positive directional selection, indicating adaptation of gene E in those species (red lineages in the tree). dN/dS < 1 suggests balancing selection, such that mutations of gene E are reducing the fitness of those species (orange through blue lineages).