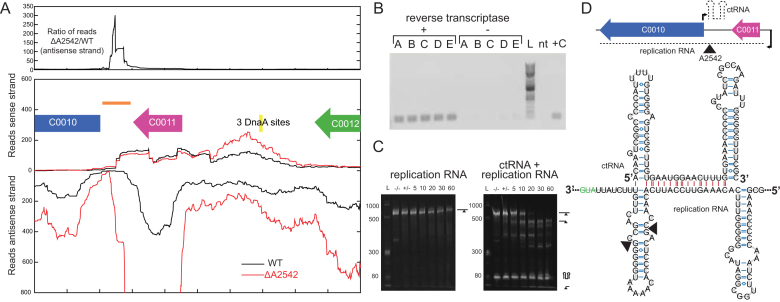
Figure 4.
Identification of ctRNA on pAQ3. (A) RNA-seq coverage of portion of pAQ3. An example replicate is shown for WT and ΔA2542 with reads aligning to the sense strand displayed at the top and reads aligning to the antisense strand displayed in the opposite direction. The top panel shows the ratio of ΔA2542 to WT of the antisense strand. (B) Verification of expression of ctRNA by amplification off of cDNA (86 bp product). Replicates A-E made from WT RNA are shown along with a reference 2 log ladder (L), a no-template (nt), and positive control (+C). (C) In vitro cleavage assays of the replication RNA and the replication RNA and ctRNA with A2542. Cleavage reactions containing 5 μg of RNA were initiated by addition of MgCl2 and samples were removed at 5, 10, 20, 30 and 60 minutes and run on a urea polyacrylamide gel. Initial samples were also taken before the addition of enzyme (–/–) and MgCl2 (+/-). (D) A diagram of the location and orientations of these two RNAs and the predicted structure of the ctRNA as predicted by mfold (38). A section of the replication RNA is shown with the marked sites of A2542 cleavage in the in vitro cleavage reaction. Intramolecular bonds are shown in blue and intermolecular interactions are shown in red. The structured RNA is shown, but these RNAs are perfectly complementary (being transcribed from the same DNA segment), and could exist as a perfectly double-stranded 86 bp region.