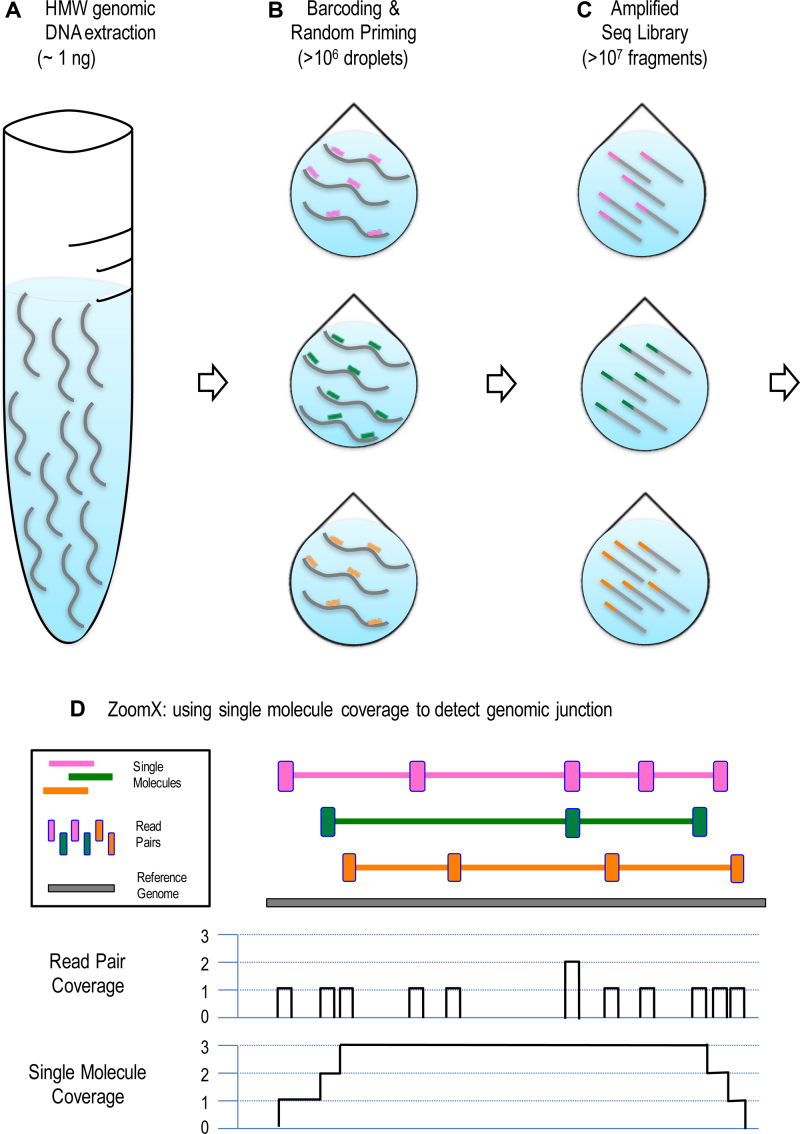
Figure 1.
Identifying rearrangements from linked reads. The workflow is illustrated in steps (A) to (D): (A) 1 nanogram of high molecule weight (HMW) DNA is extracted from the sample; (B) the extraction was partitioned to > 106 droplets, where in average only a few DNA molecules enter each droplet and get the same barcode; The barcode, uniquely colored, is linked to random primers which sparsely prime on the HMW DNA; (C) the primed DNA undergoes several rounds of displacement amplification to generate short fragment within the droplet, which will be released into one sequencing library pool; (D) the linked-read sequencing is performed and ZoomX infers single molecules based on aligned barcode linked reads; ZoomX scans genome coordinate pairs to detect if there is any rearrangement junction in between based on single molecule coverage. In the plot: Each DNA molecule (fragment) is represented by a gray curved (linear) segment; each color represents a unique barcode. In (B); each short segment represents a random primer with barcode. In (D): Each colored long stretch is an inferred linked-read molecule; Each colored vertical short bar is a linked-read pair, which is interspersed along the inferred molecule; We depict the single molecule coverage against the base pair coverage given the shown molecules. The single molecule coverage is higher and more consistent across the genome.