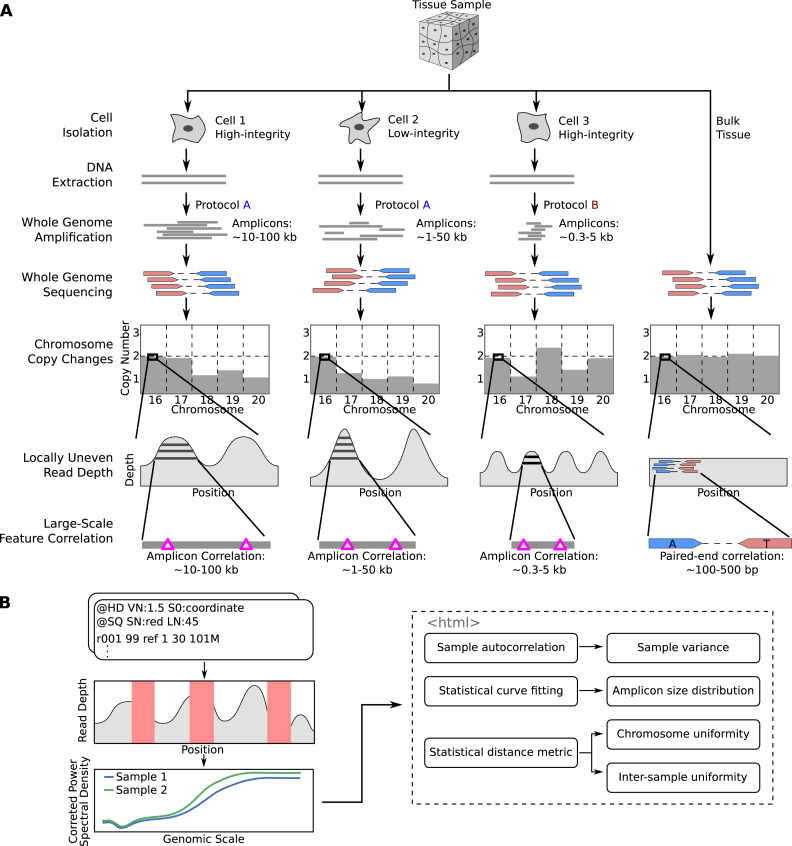
Figure 1.
Overview of single cell whole-genome sequencing and sources of artefacts, and the PaSD-qc pipeline. (A) Schematic overview of single-cell whole genome sequencing and the artefacts created by whole-genome amplification. The extent and patterns of the biases depend on the cell condition (high- or low-integrity) and on the scWGS protocol used (protocol A or protocol B). The pink triangles in the ‘Large-Scale Feature Correlation’ represent genomic events (e.g., single nucleotide variants) which are spanned by a single amplicon and are thus correlated. The only correlation pattern present in bulk sequencing is due to paired-end sequencing, represented by positions marked ‘A’ and ‘T’ spanned by the mate pair. (B) Schematic overview of the PaSD-qc pipeline. Read depth is extracted from bam files at uniquely mappable positions. Red rectangles represent regions where the true read depth is unknown due to low mappability, locus dropout, or sequencing bias. PaSD-qc uses a custom power spectral density estimation procedure to accurately estimate the correlation patterns in the data, and these patterns are then used to assess amplification properties and quality control measures. By default, the results are summarized in an interactive HTML report.