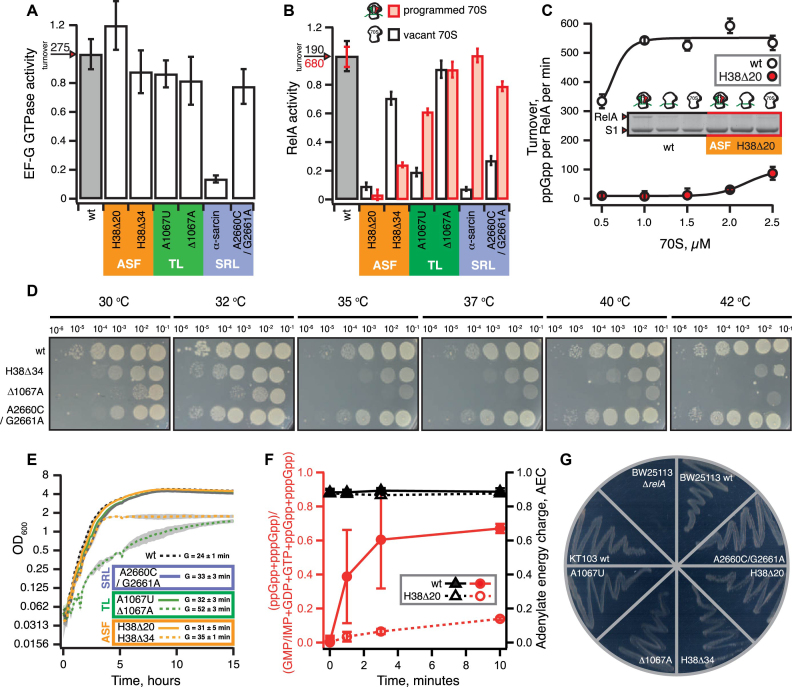
Figure 4.
Effects of molecular perturbations of ASF, SRL and TL on RelA’s enzymatic activity. GTPase activity of 0.1 μM EF-G assayed in the presence of 0.3 mM of 3H GTP and 0.5 μM 70S (A). 3H-ppGpp synthesis by 30 nM RelA assayed in the presence of either 1 mM ATP and 0.3 mM 3H GDP as well as either 70S ribosomes (0.5 μM; black bars) or ‘starved’ ribosomal complexes (0.5 μM 70S, 2 μM mRNA(MF), tRNAiMet and tRNAPhe, 2 μM each; red bars) (B). 3H-ppGpp synthesis by RelA in the presence of increasing concentrations of either wild type (empty circles) or H38Δ20 (red filled circles) 70S programmed with 2 μM mRNA(MF) as well as tRNAiMet and tRNAPhe, 2 μM each (C). Association 1 μM RelA with either 1 μM wild type or H38Δ20 70S ribosomes programmed with 2 μM mRNA(MF) as well as tRNAiMet and tRNAPhe, 2 μM each (C, insert). Growth of Δ7 E. coli KT101-based strains expressing either wild type or mutant 23S rRNA from pRB103 plasmid on LB plates at varied temperature (D), in the liquid LB culture at 37°C (E) and on SMG plates at 37°C (G). Serial dilution spotting analysis on LB plates was scored at 12 (35°C, 37°C, 40°C and 42°C) or 21 (30°C and 32°C) hours of incubation (D). Nucleotide pools in Δ7 E. coli KT101-based strains expressing either wild type (filled symbols) or H38Δ20 mutant (empty symbols) 23S rRNA from pRB103 plasmid grown on MOPS liquid media supplemented with 0.4% glucose and challenged by mupirocin added to final concentration of 150 μg/ml (F). The error bars represent SD of the mean of biological replicates (n = 3). All biochemical experiments were performed at 37°C in HEPES:Polymix buffer, pH 7.5 at 5 mM Mg2+. Uninhibited turnover values corresponding to 1.0 activity in the presence of wild type ribosomes are provided on individual panels A and B. Error bars represent SDs of the turnover estimates by linear regression if not stated otherwise. Each experiment was performed at least three times.