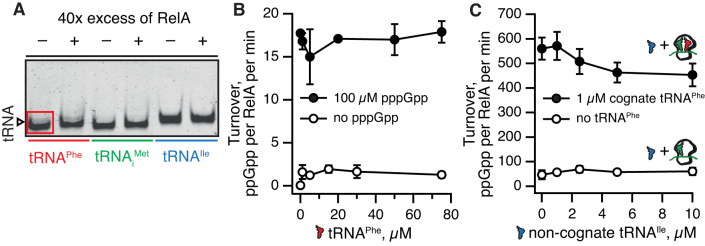
Figure 5.
RelA does not form a stable complex with deacylated tRNA off the ribosome. No RelA:tRNA complex with either E. coli tRNAPhe, tRNAiMet or tRNAIle is detected by EMSA, as judged by the absence of an extra band on the gel in the presence of RelA (A). Deacylated tRNAPhe does not activate the enzymatic activity of 250 nM RelA either in the absence (empty black circles) or presence (filled black circles) of 100 μM pppGpp (B). Non-cognate deacylated tRNAIle neither significantly inhibits ppGpp synthesis by 50 nM RelA in the presence of ‘starved’ ribosomal complexes (filled black circles) nor activates RelA in the presence of ribosomal complexes with a vacant A-site (empty black circles) (C). EMSA assays were performed in the presence of 70 nM tRNA and 2.8 μM RelA (A). 3H-ppGpp synthesis by RelA was assayed in the presence of 1 mM ATP and 0.3 mM 3H GDP as well as increasing concentrations of deacylated tRNA as indicated on the figure (B and C). Ribosomal complexes were formed in situ by combining 0.5 μM 70S, 1 μM mRNA(MF) and 1 μM tRNAiMet either in the presence or absence of 1 μM cognate A-site tRNAPhe (C). Experiments with ribosomal complexes were performed in the presence of 100 μM ppGpp. All experiments were performed at 37°C in HEPES:Polymix buffer, 5 mM Mg2+, pH 7.5. Error bars represent SDs of the turnover estimates by linear regression and each experiment was performed at least three times.