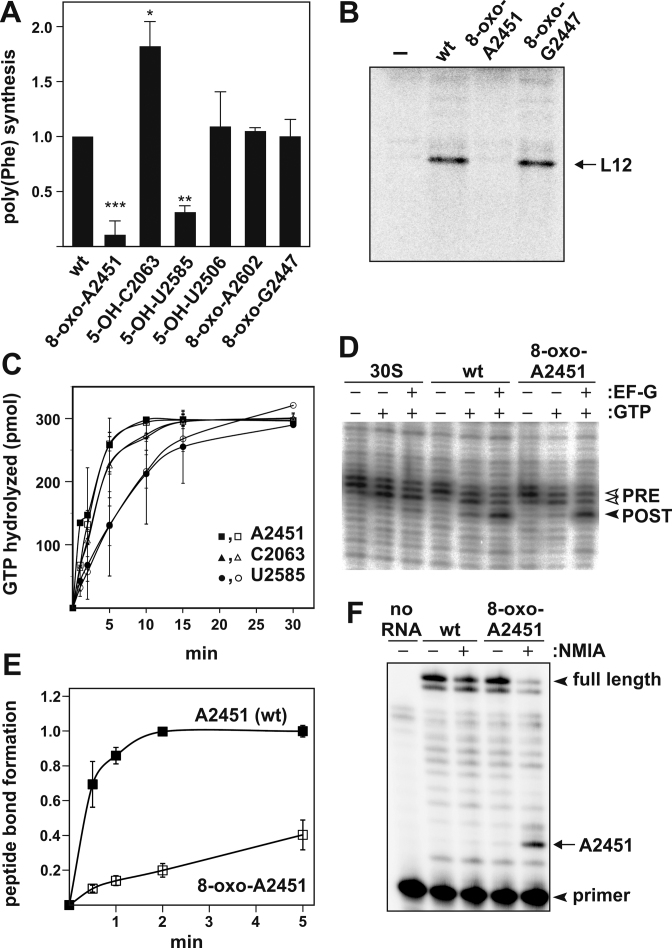
Figure 2.
Oxidation in the ribosome's active site differentially affects protein synthesis. (A) Endpoint measurements in poly(Phe) synthesis employing in vitro reconstituted ribosomes containing either synthetic RNA oligonucleotides with the wildtype sequence (wt) in the active site, or oligos harboring specific nucleobase oxidations in the PTC or nearby regions. In all reconstituted cp-23S rRNA constructs the minute translational activities of ribosomes containing no oligo was subtracted as background, and the activity of ribosomes containing the corresponding wt oligonucleotide was taken as 1.00. Values represent the mean of at least three independent experiments, with SD shown as error bars. Significance according to paired Student's t-test: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. (B) Representative autoradiogram of an SDS-PAGE showing in vitro translation of L12 r-protein mRNA by reconstituted ribosomes carrying 8-oxo-adenosine at 23S rRNA position A2451 or 8-oxo-guanosine G2447. Reactions with ribosomes reconstituted without a complementing synthetic RNA oligo (–) served as negative control. (C) EF-G GTPase activation as measured by the rate of GTP hydrolysis using reconstituted ribosomes with the wt sequence (filled symbols) or oxidized PTC bases (open symbols). Values represent the mean of at least two independent time course experiments, with error bars indicating SD. (D) Translocation of ribosomes oxidized at position A2451 along a heteropolymeric mRNA as determined by toeprinting. Toeprint bands indicate the positions of mRNA-bound ribosome complexes before (PRE) and after (POST) addition of EF-G and GTP. Ribosomal complexes carried deacylated tRNA and peptidyl-tRNA in the P- and A-site in the PRE state, and in the E- and P-site in the POST complexes, respectively. Reactions in the presence of T. aquaticus 30S subunits alone served as negative controls. (E) Time course of peptide bond formation in ribosomes containing either the wt sequence or 8-oxo-A2451. Values represent the mean of 3 independent time course experiments. The activities of in vitro assembled ribosomes without a synthetic oligo were subtracted as background. The endpoint value of ribosomes containing the wt sequence was taken as 1.00. (F) SHAPE probing of the 8-oxo-A2451 oligo inside the reconstituted large ribosomal subunit. Reconstituted 50S were treated with NMIA to determine ribose 2′-OH reactivity, which was detected by primer extension analysis. The location of the radiolabeled primer, the full length reverse transcription product and the position of A2451 in the denaturing polyacrylamide gel are indicated by arrow heads and an arrow, respectively. Note that the NMIA reaction product halts reverse transcription one position 3′ of the reacted site. Primer extension reaction in the absence of any RNA was used as negative control (no RNA).