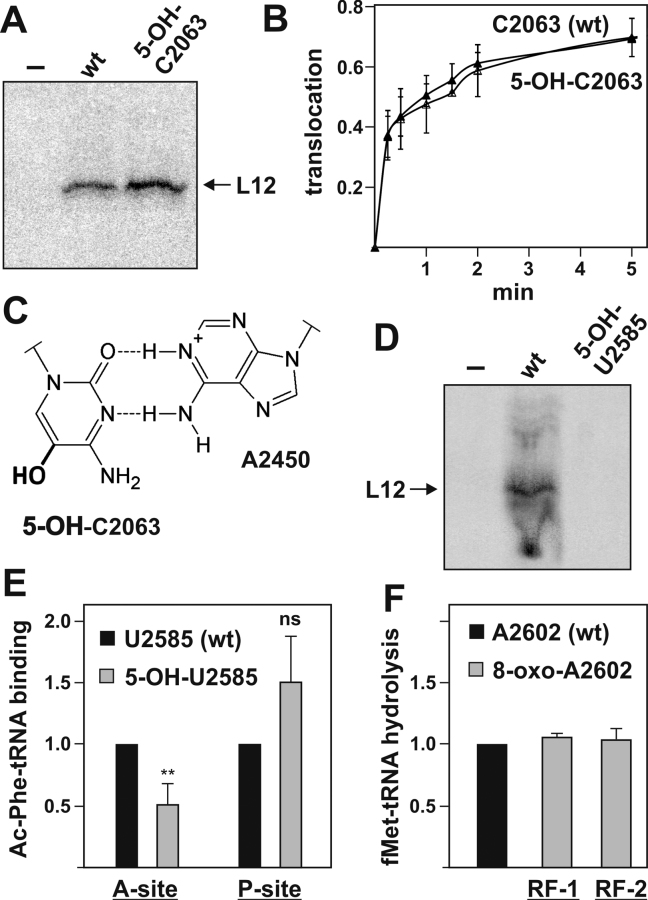
Figure 3.
Effects of oxidation at positions C2063, U2585 and A2602. (A) In vitro translation of r-protein L12 mRNA with chemically engineered ribosomes oxidized at position C2063. Reaction with ribosomes reconstituted without a complementing synthetic RNA oligo (–) served as negative control. (B) Time course of translocation of ribosomes oxidized at position C2063 along a heteroplymeric mRNA as determined by toeprinting. The translocation reaction was initiated by the addition of EF-G•GTP and stopped by the antibiotic neomycin at the indicated time points. The mean and standard deviation of three independent time course experiments are shown. (C) The non-Watson-Crick base pair formed in the 50S subunit between C2063 and A2450 is shown here with 5-OH-C (bold) in position 2063. The integrity of this interaction was shown previously to be vital for effective translation (40). (D) In vitro translation of r-protein L12 mRNA with chemically engineered ribosomes oxidized at position U2585. Reaction with ribosomes reconstituted without a complementing synthetic RNA oligo (–) served as negative control. (E) Ac-[3H]Phe-tRNAPhe binding affinity to the A- or P-site, respectively, in chemically engineered ribosomes oxidized at position U2585. Binding activities were normalized to ribosomes harboring the synthetic wt oligo, whereas unspecific tRNA binding to ribosomes containing no synthetic oligo was always subtracted as background. The mean and standard deviation of 5–7 independent binding experiments are shown. Paired Student's t-test: **P ≤ 0.01; ns, statistically not significant. (F) Release reaction as measured by P-site-bound f[3H]Met-tRNAfMet hydrolysis after RF-1 (n = 2) or RF-2 (n = 4) addition to chemically engineered ribosomes carrying an 8-oxo-adenosine at position A2602. f[3H]Met-tRNAfMet hydrolysis was normalized to wt activities and f[3H]Met release in ribosomes containing no oligo was subtracted as background.