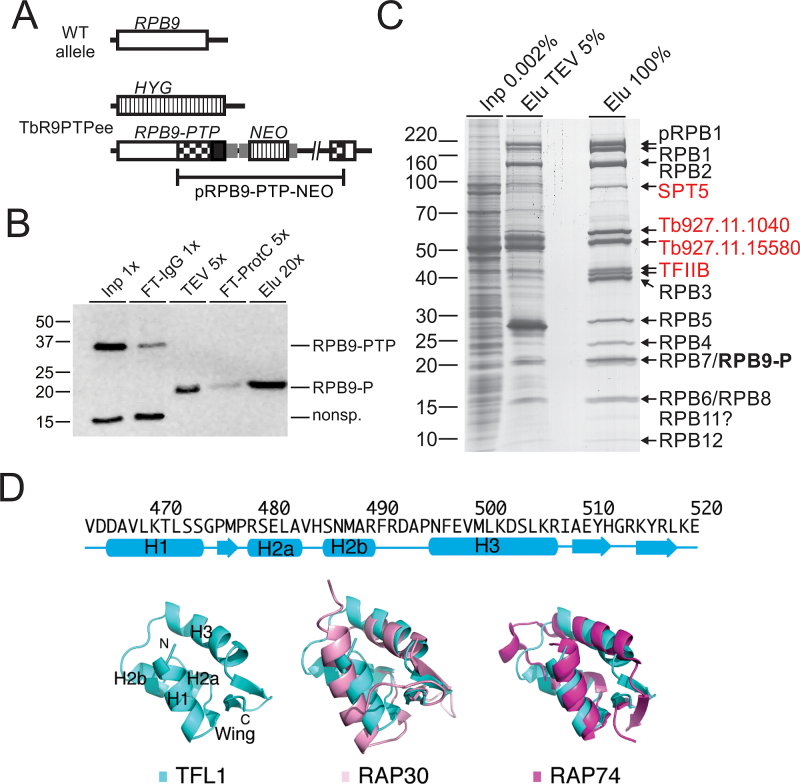
Figure 1.
Tandem affinity purification of RNA pol II via RPB9-PTP. (A) Schematic depiction (not to scale) of a wild-type RPB9 allele (open box) and the two modified RPB9 alleles in TbR9PTPee trypanosomes. In this cell line, one RPB9 allele was knocked out by a hygromycin phosphotransferase gene (HYG, striped box) and integration of plasmid RPB9-PTP-NEO into the second allele fused the PTP sequence (black box) to the RPB9 coding region and added a neomycin phosphotransferase gene (NEO; striped box). Smaller gray boxes indicate gene flanks for RNA processing signals and checkered boxes depict RPB9 sequences encoded in the plasmid. (B) Immunoblot monitoring of RPB9-PTP tandem affinity purification, detecting the tagged protein with the monoclonal anti-ProtC antibody HPC4 in extract (Inp), flowthrough of IgG affinity chromatography (FT-IgG), TEV protease eluate, flowthrough of anti-ProtC immunoaffinity chromatography (FT-ProtC) and the final eluate (Elu). x-Values indicate relative amounts analyzed (Please note that TEV protease, cleaving off ProtA, reduces RPB9-PTP to RPB9-P, which corresponds to molecular mass reduction of ∼20 kDa). Marker sizes in kDa are indicated on the left. (C) RPB9-PTP co-purified proteins (Elu) and, for comparison, proteins of extract (Inp) and the TEV eluate were separated on a 10–20% SDS polyacrylamide gradient gel and stained with Coomassie blue. Individual protein bands were excised and proteins identified by LC/MS/MS as indicated on the right. (D) Predicted secondary-structure elements and homology model for the C-terminal winged-helix DNA-binding motif of TFL1. Top, secondary-structure assignments predicted by PHYRE 2 are shown as cyan cylinders (α helices) and arrows (β strands). Bottom, ribbon diagram of top scoring PHYRE homology model of the C-terminal domain of TFL1 (left), and supposition of TFL1 (cyan) with the C-terminal WH DNA-binding domain of human RAP30 (PDB ID: 2BBY) (pink; middle) and RAP74 (PDB ID: 1I27) (magenta; right).