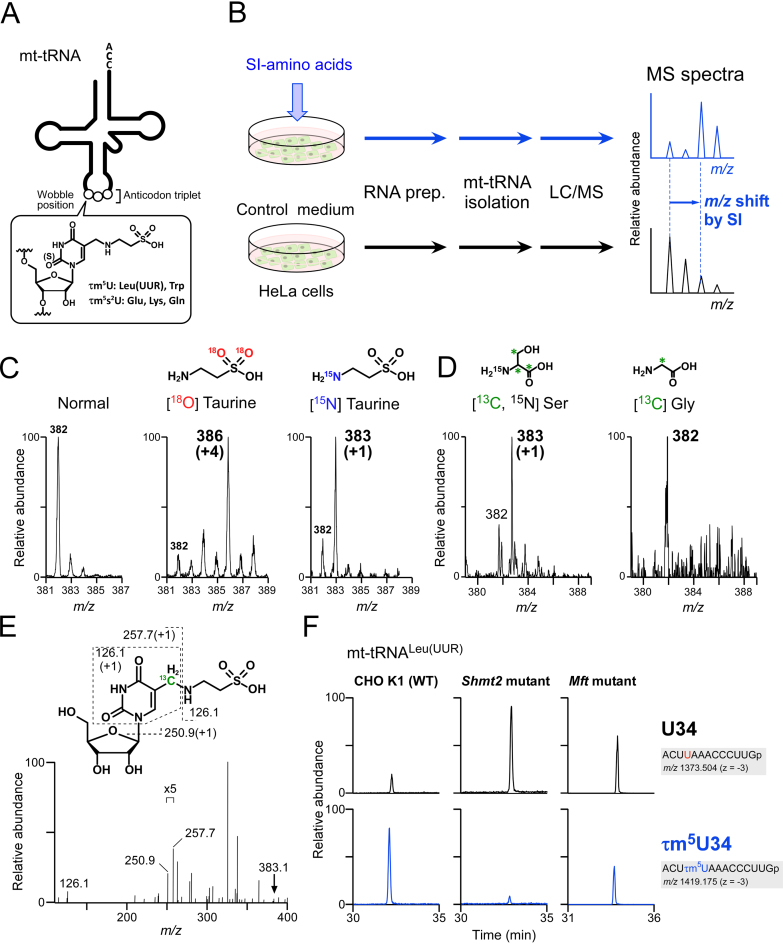
Figure 1.
Metabolic labeling of mt-tRNAs to determine metabolic sources of τm5U. (A) Chemical structure of τm5(s2)U which is present at the wobble (first) position of anticodon of five mt-tRNAs. (B) Scheme for metabolic labeling experiments. HeLa cells were cultured with stable isotope (SI)-labeled or non-labeled amino acids. Total RNA was extracted from each culture, and individual mt-tRNAs were isolated and digested into nucleosides, followed by liquid chromatography–mass spectrometry (LC/MS) analysis to detect SI-labeled τm5(s2)U. (C) Mass spectra of τm5U in mt-tRNATrp isolated from HeLa cells cultured in normal medium (first from the left), [18O] taurine–supplemented medium (second) and [15N] taurine–supplemented medium (third). Labeled atoms are colored as indicated. Increased m/z values of τm5U due to the presence of SI-atoms are shown in parentheses. (D) Mass spectra of τm5U in mt-tRNAs for Trp and Leu(UUR) isolated from HeLa cells cultured in [15N, 13C3] Ser–supplemented medium (left) and [13C] Gly–supplemented medium (right). 13C atoms in their chemical structures are indicated by asterisks. (E) Collision-induced dissociation (CID) spectrum of [13C]-labeled τm5U nucleoside. The precursor ion (m/z 383.1) for CID is marked by an arrow. Product ions in the CID spectrum are assigned in the chemical structure of τm5U. (F) Mitochondrial 1C metabolism involved in τm5U biosynthesis (see Figure 7). Extracted mass chromatograms (XICs) of anticodon-containing RNase T1-digested fragments bearing U34 (upper panels) or τm5U34 (lower panels) from mt tRNALeu(UUR) isolated from Chinese hamster ovary (CHO) wild-type (WT) (left panels), Shmt2 mutant (center panels) and Mft mutant (right panels) cells. Sequence of the detected fragment, with its m/z value and charge state, is indicated on the right.