Abstract
The role of DNA sequence in determining replication timing (RT) and chromatin higher order organization remains elusive. To address this question, we have developed an extra-chromosomal replication system (E-BACs) consisting of ∼200 kb human bacterial artificial chromosomes (BACs) modified with Epstein-Barr virus (EBV) stable segregation elements. E-BACs were stably maintained as autonomous mini-chromosomes in EBNA1-expressing HeLa or human induced pluripotent stem cells (hiPSCs) and established distinct RT patterns. An E-BAC harboring an early replicating chromosomal region replicated early during S phase, while E-BACs derived from RT transition regions (TTRs) and late replicating regions replicated in mid to late S phase. Analysis of E-BAC interactions with cellular chromatin (4C-seq) revealed that the early replicating E-BAC interacted broadly throughout the genome and preferentially with the early replicating compartment of the nucleus. In contrast, mid- to late-replicating E-BACs interacted with more specific late replicating chromosomal segments, some of which were shared between different E-BACs. Together, we describe a versatile system in which to study the structure and function of chromosomal segments that are stably maintained separately from the influence of cellular chromosome context.
INTRODUCTION
Mammalian DNA replication is temporally and spatially regulated in units termed replication domains (RDs) (1). RDs are 400–800 kb segments of coordinately regulated replication timing (RT) change during cell differentiation (2), and correspond to chromatin structural units defined by chromatin conformation capture as Topologically Associated Domains (TADs) (3–5). Importantly, early and late replicating RDs form two distinct self-interacting compartments inside the nucleus, A and B, such that chromatin that replicates at similar times tends to be in closer proximity in the nucleus (6). A/B compartments occupy distinct positions in the interior or periphery of the nucleus, respectively (7,8), consistent with earlier cytogenetic observations of the distribution of pulse labeled DNA synthesis known as replication foci (9). Functionally, early and late replicating RDs correlate in a general sense with active and inactive chromatin, respectively, and changes in RT during cell fate transitions often coincide with changes in transcriptional activity of the resident genes, although the relationship is complex (10). Early and late replicating regions also exhibit distinct patterns of mutations (11,12), and disruptions in RT appear in several diseases (13,14), which may be linked to aberrant gene expression or genome instability.
Several lines of evidence suggest the importance of DNA sequence in regulating RT, compartmentalization and related large-scale chromatin properties. For example, human chromosome 21 retains a human-like RT pattern when introduced into a transgenic Down syndrome mouse model (15). Tandem integration of multiple BACs into the genome results in their self-assembly into domains of active or repressive chromatin marks with transcriptionally active regions decorated with H3K27me3 located at the exterior and H3K9me3 and HP1 enriched in the interior (16). In addition, a recent study characterizing the spatial arrangement of a 4.26 Mb human artificial chromosome (HAC) (17) in a xenospecific mouse background demonstrated sequence specific conformation of the HAC using 4C and FISH (18). Specifically, gene-rich and SINE-rich euchromatin, gene-poor and LINE/LTR-rich heterochromatin, and gene-depleted and satellite DNA-containing constitutive heterochromatin on the HAC interacted preferentially with chromatin of similar properties in the genome. However, these general sequence properties are not sufficient to dictate RT and cannot explain RT changes during development (1). Hence, the role of DNA sequences in determining large-scale chromatin structure and function and its regulation during development remain challenging questions. Here, we adopted an autonomously replicating system to test the sufficiency of isolated sequences in determining RT and sub-nuclear compartment without interference from integration.
Epstein-Barr Viruses (EBV) is a chronic intra-cellular parasite in infected individuals, stably maintaining its circular 165 kb genome extra-chromosomally in host cells during latency (19,20). A 1.8 kb region of the EBV genome containing a replication origin termed OriP, which is composed of Dyad Symmetry (DS) and Family of Repeats (FR) elements, and can support stable retention of circular plasmids in human cells (20–23). The ability of EBV to stably maintain its circular genome is granted by the FR element, in coordination with the viral protein, Epstein-Barr Virus Nucleus Antigen 1 (EBNA1) (24,25). EBNA1 binds to the FR during mitosis in a fashion that permits FR containing plasmids to be transmitted approximately equally to daughter cells (26). The other component of EBV OriP, the DS element, functions as a replication origin in small circular plasmids, but in the full length EBV genome replication initiation does not occur at the DS but instead occurs at many sites distributed throughout the EBV genome (27–30), and, in fact, the DS is dispensable for full-length EBV replication (31,32). A previous study has shown that any DNA longer than ∼10 kb harbored on an FR-containing plasmid and introduced into an EBNA1-expressing human cell line can replicate efficiently and autonomously (33), and that replication proceeds semi-conservatively, once per cell cycle and initiates at multiple sites, including within prokaryotic DNA segments (34,35). In one study, three plasmids carrying 14–20 kb fragments of human DNA were found to replicate at specific times during S phase, but not necessarily at the same time as in the chromosome (22). It was speculated that the segments may not have been long enough to establish proper RT. Later work showed that Bacterial Artificial Chromosomes (BACs) containing the FR can be stably retained in EBNA1-expressing human or mouse cells, thus this system was proposed as a potential gene therapy application for transgene expression (36–39). In fact, a beta-globin gene carried by this vector system was reported to be transcribed and correctly spliced in murine fibroblasts (37). Therefore, we sought to investigate whether these extra-chromosomal autonomous replicating units could serve as useful tools to dissect the role of DNA sequences in RT regulation, avoiding interference from neighboring regions that is experienced with ectopic integration approaches. Our results show that ∼200 kb segments can establish RT and sub-nuclear compartment de novo on E-BAC vectors.
MATERIALS AND METHODS
Cell culture
HeLa S3 cells were cultured in 10% cosmic calf serum supplemented Dulbecco's Modified Eagle Medium (DMEM). HeLa-EBNA1 and E-BAC transfected cells were grown in 500 mg/ml G418 and 0.2 mg/ml hygromycin B, respectively. Human iPSC K3 or K3-EBNA1 were grown in StemPro (Thermo Fisher, A100701) culture media on Geltrex (Thermo Fisher, A14133) coated dishes. HeLa-EBNA1 and K3-EBNA1 was made by electroporating AttI linearized p2091 plasmid (gift from Bill Sugden lab). One G418 resistant colony was expanded for the study, and K3-EBNA1 was maintained under 50 ug/ml G418 selection.
Recombineering and E-BAC transfection
FR cassette was prepared by digesting pDY-HA1-HA2-neo plasmid with BsiWI and EcoRV. Plasmid pDY-HA1-HA2-neo was made by replacing AmpR gene at AseI-SspI sites with HA1-neoR fragment and inserting HA2 fragment into NruI site of pDY-plasmid (gift from Michele P. Calos Lab). The two homology arms (HAs) are 527 and 535 bp sequences from the sacB gene on pBACe3.6 vector. BACs were ordered from BACPAC Resource Center (www.bacpac.chori.org/), fingerprinted using restriction mapping, and electroporated to sw105 [(cro-bioA) <> araC-PBADflpe] using Bio-Rad Gene Pulser using conditions of 1.2 kV, 25 μF and 200 Ω. Sw105 carrying BACs were inoculated at 42°C for exactly 15 min to induce the expression of exo, bet, and gam recombination proteins, after which competent cells were prepared and FR cassette was introduced by electroporation. Cells were grown on low-salt LB broth with 25 ug/ml kanamycin at 32°C. E-BACs were confirmed by colony PCR (Supplementary methods), purified using Qiagen large-construct kit, and 2 ug was transfected to HeLa-EBNA1 using Qiagen effectene according to the manual. pDY-HA1-HA2-neo-GFP plasmid was constructed by replacing CAG-GFP with sequences between Avr2 and mfe1 site of pDY-HA1-HA2-neo. FR-GFP cassette was prepared by digesting pDY-HA1-HA2-neo-GFP with SspI and EcoRV. Sox17-1 GFP E-BAC was transfected into K3-EBNA1 cells by nucleofection according to manufacture's specifications (Lonza P3 Primary Cell Kit, V4XP-3032). GFP positive cells were enriched by FACS, and expanded before performing Gardella gel and repli-seq.
Gardella gel and episome isolation
Horizontal Gardella gel was prepared based on (40). Cells were resuspended in sample buffer A (15% Ficoll, 40ug/ml RNaseA, 0.01% bromophenol blue in TBE), loaded in each well, and laid over by lysis buffer (5% Ficoll, 1% SDS, freshly added 1mg/ml pronase, Roche 10165921001, 0.05% xylene cyanol in TBE). Electrophoresis was carried out at 35 V for 5 h, followed by 109 V for 32.5 h in 1× TBE (Tris–borate–EDTA buffer). DNA was fragmented by staining with 1 ug/ml ethidium bromide for exactly 30 min with constant agitation, followed by UV irradiation at 60 mJ of energy (UVP CL-1000 UV crosslinker). DNA was then denatured in 0.4 N NaOH, 1.5 M NaCl for 15 min, neutralized using 87.66 g of NaCl, 60.5 g Tris–base, 1 L ddH2O, pH 7.5, and transferred to GE healthcare Amersham Hybond–N+ membrane overnight in Capillary blotting apparatus in 20× SSC (300 mM Tri-Na-citrate, 3 M 175.32 g NaCl, pH 7). DNA was covalently linked to the membrane by UV radiation. After pre-hybridized at 65°C with gentle agitation for 30 min, the membrane is hybridized in Church and Gilbert buffer at 65°C overnight, followed by stringency Wash in following conditions: 2× SSC, 0.1% SDS for 5 min at RT, twice; 1× SSC, 0.1% SDS for 15 min at RT; and 0.1× SSC, 0.1% SDS for 10 min at 65°C, twice. Probe was P32 dATP labeled PCR fragment from pBACe3.6 (F: 5′-tgctggcttttcaagttcct, R: 5′-gggcaccaataactgcctta) using random priming DNA labeling system (Life Technologies 18187-013). Lastly, signals were detected using Typhoon imaging system.
Genome-wide replication timing profiling
Genome-wide RT profiles were constructed as previously described (45,46). Briefly, cells were pulse labeled with BrdU for 2 h and separated into early and late S-phase fractions by Propidium iodide staining and flow cytometry. BrdU-substituted DNA from 40k early and late S-phase cells was immunoprecipitated by anti-BrdU antibody (Becton Dickinson 347580), and sequencing libraries were prepared by NEBNext Ultra DNA Library Prep Kit for Illumina (E7370). Sequencing was performed on Illumina HiSeq 2500 using 50 bp single end. Reads of quality scores above 30 were mapped to customized hg19 reference genome with BAC backbone using bowtie2. Read counts were binned into 6 kb non-overlapping windows, and log2 ratio of read-counts between early and late fraction are plotted in R. All profiles were quantile normalized to yield the same density distribution.
E-BAC copy number
E-BAC copy numbers were calculated using the following formula using read numbers in repli-seq data:
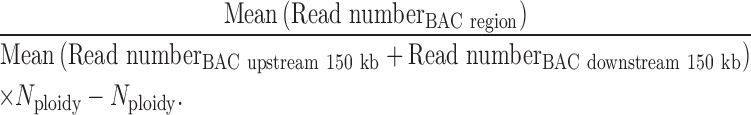 |
N ploidy refers to the copy number of BAC corresponding chromosomal regions. Since the HeLa genome has a high degree of copy number variation (CNV) we used a combination of our total repli-seq reads and prior quantification of CNV in HeLa cells (41) to determine copy number. For Sox17-3 and Sox17-1 BAC, Nploidy = 3; for BAC5, Nploidy = 4. We also compensated for the deletion in BAC5 clone 1 and clone 2 by removing the deleted region from the mean (read number BAC region). Nploidy for K3-EBNA1 cells is two.
4C-seq
4C-seq was performed as previously described (42) with minor modifications. 10 million cells were fixed in 1% formaldehyde, and chromatin was digested using HindIII before proximity ligation, followed by DpnII after reverse crosslinking. 4C libraries were prepared using inverse-PCR primers that contain illumina adapters and anneal to the pBACe3.6 bacterial sequences (forward primer: 5′-ATAAACTACCGCATTAAAGC; reverse primer: 5′-GGAGCCACTATCGACTACGC). Libraries were sequenced with 3–7 million mappable reads using 50 bp single-end sequencing with Illumina HiSeq 2500. Reads were mapped to hg19 build using BWA. Each profile was then normalized to RPM, binned to 10 kb windows, and smoothed using rolling mean smoothing across 15 windows. 4C-seq strong interaction sites (SIs, 39-400RPM) or moderate interactions sites (MIs, 20-39RPM) are considered shared between two datasets if they overlap by 20 kb upstream or downstream. Shared sites were counted using bedtools Intersect function.
RESULTS
Characterization of the E-BAC system
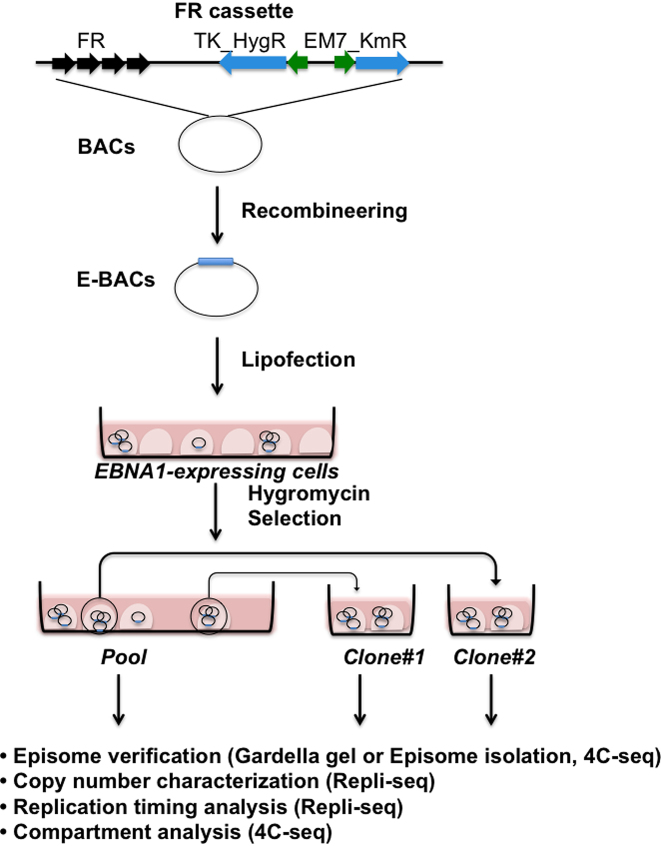
Figure 1 illustrates the E-BAC system. HeLa cells stably expressing EBV EBNA1 were transfected with the E-BACs harboring an FR cassette inserted by recombineering into the BACs listed in Figure 2A (and Supplementary Figures S1 and S2). Hygromycin resistant colonies were collected as well as the pool of remaining resistant cells. Stable extra-chromosomal replication of the E-BACs was confirmed by Gardella gel (40) or Hirt extraction (43). Gardella gel loads live cells directly into gel electrophoresis wells and lyses them in situ, while Hirt extraction isolates E-BACs from transfected HeLa-EBNA1 cells by alkaline lysis. In both methods, 200kb BAC DNA, but not mammalian chromosomes, can enter the gel during electrophoresis, as detected by Southern hybridization. Finally, copy number and RT of the E-BACs were measured by repli-seq (Marchal et al. accepted), and chromatin compartments were evaluated by 4C-seq (42) using a bait within the pBAC3.6 plasmid vector sequences to assay genome-wide interaction partners (Figure 1). 4C is also a very sensitive detection method for integration events, since huge numbers of 4C interaction partners will derive from sequences close to any integration site.
Figure 1.
E-BAC experimental design. FR cassette contains the EBV family of repeats (FR) and selectable markers for recombineering in E. coli (Em7_KmR) and for selection in mammalian cells (TK_HygR). The FR cassette is flanked by sequences homologous to the pBAC3.6 vector sequences, and inserted into candidate BACs by recombineering (see Materials and Methods). The resulting E-BAC is then transfected into a cell line stably expressing the EBV EBNA1 protein (example: HeLa-EBNA1). After selection for the E-BAC, drug resistant colonies were collected as well as the pool of remaining resistant cells, which are analyzed as indicated for autonomous replication and copy number before repli-seq and 4C-seq are performed.
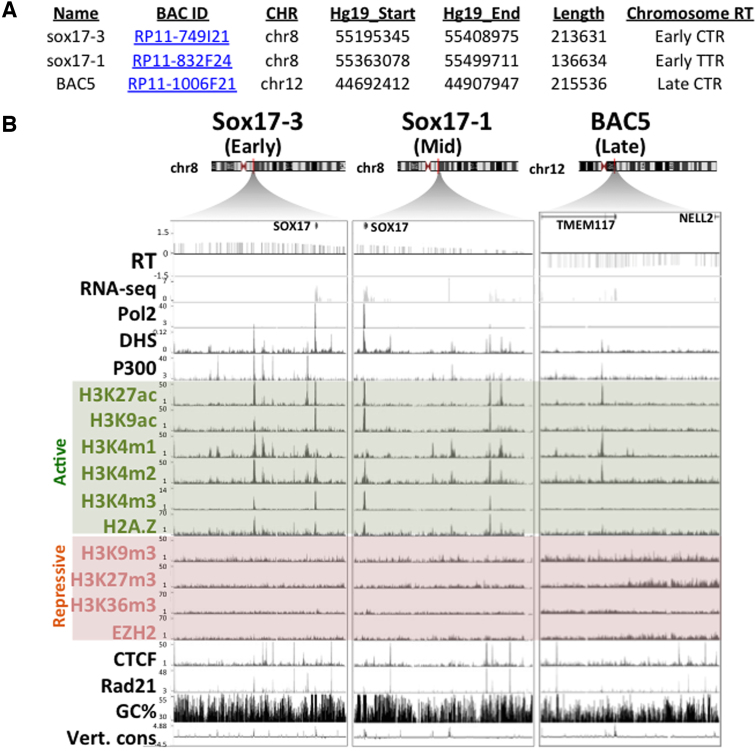
Figure 2.
Candidate BACs used in this study. (A) Summary of candidate BACs. (B) UCSC genome browser tracks demonstrating gene position (Gene), replication timing (RT), DNase I hypersensitive sites (DHS), chromatin immunoprecipitation with antibodies to RNA polymerase 2 (Pol2), histone post-translational modifications and variants, polycomb PRC2 subunit EZH2, architectural protein CTCF, cohesin subunit Rad21, and degree of vertebrate conservation (Vert. cons). Active and repressive histone marks are shaded in green and red, respectively.
Figure 2 summarizes the three chromosomal segments contained within the BACs chosen to represent an early replicating constant timing region (CTR), the early replicating border of the Timing transition region (TTR) adjacent to the same CTR, and a late CTR (Figure 2) (44). In repli-seq data CTRs are interpreted to be domains of coordinate activation of multiple adjacent replicons, activated stochastically so that different cells are using different cohorts of origins, which averages out to a region of relatively constant replication timing in the population repli-seq data (45). TTRs are regions of suppressed origin activity, where forks travel mostly unidirectionally for many hours (45). The sox17-3 BAC corresponds to a segment within an early CTR on chromosome 8 that contains the gene sox17 (Figure 2B), which is actively transcribed in HeLa cells. The native locus in HeLa is depleted of repressive histone marks, with two active promoter/enhancer sites including the Sox17 promoter, DNaseI hypersensitive site (DHS) peaks and active histone marks H3K27ac and p300. Therefore, the sox17-3 BAC region represents a typical early replicating region, with DNase hyper-sensitive sites (DHSs) associated with active or poised regulatory elements (Figure 2B). The sox17-1 E-BAC is derived from TTR sequences overlapping 45.9kb of the sox17-3 E-BAC including the sox17 gene, and has a similar chromatin composition to the sox17-3 region but lacks prominent enhancer (p300) sites (Figure 2B). Lastly, E-BAC BAC5 corresponds to a constitutively late replicating region on chromosome 12 that carries the 3′ and 5′ end, respectively, of two inactive genes: TMEM117 and NELL2. Compared to the sox17-1 and sox17-3 regions, the endogenous chromatin represented by BAC5 is depleted of active marks and enriched with repressive histone marks, such as H3K9m3 and H3K27m3, which may be required for targeting to the nuclear periphery (46). The AT percentages (47) of the three BACs vary slightly (58%-62%), and architectural protein binding sites such as CTCF and Rad21 are present (Figure 2).
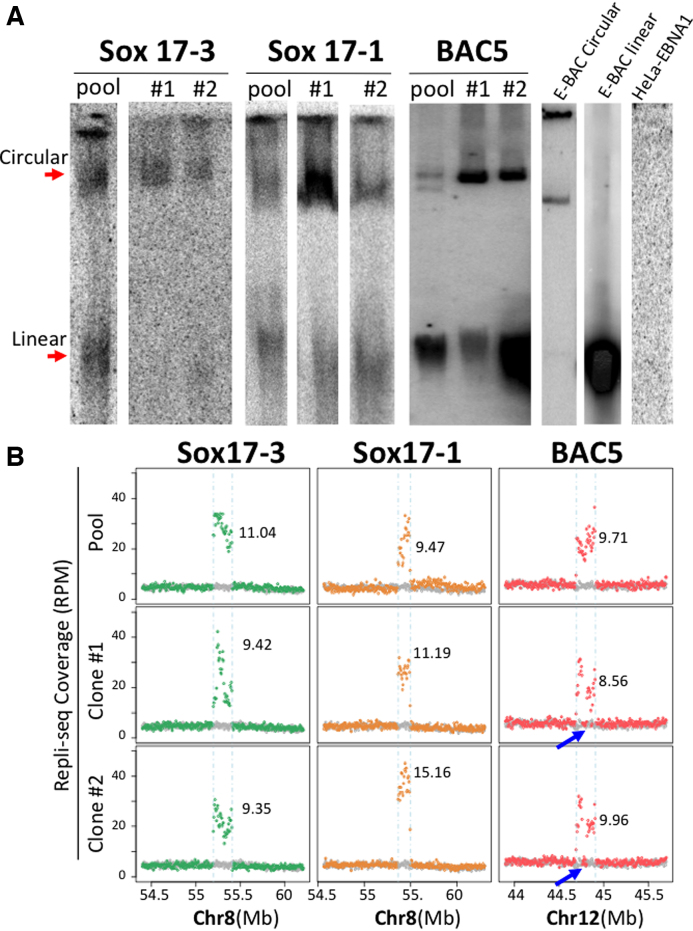
Either Gardella gel or a modified Hirt extraction detected episomal signals for all three BACs in both the transfected pools and clones, while E-BAC-free HeLa-EBNA1 cells served as a negative control (Figure 3A). Both episomal detection methods revealed similar results (Supplementary Figure S3A). Additional clones picked from the sox17-3 and sox17-1 E-BAC pool indicated that episome signals can be detected in 60–80% of clones (Supplementary Figure S3B). This number is likely to be an underestimate, as we may not detect E-BACs in clones with low E-BAC copy number. E-BAC copy number was inferred from genome-wide RT (Repli-seq; discussed below) data by comparing the read coverage differences between the BAC sequences and the neighboring regions (Figure 3B and methods). Copy numbers of three E-BACs range from 6 to 15 copies per cell on average for a given pool or subclone. The sequencing results also revealed a high degree of variation in the read coverage for E-BACs, suggesting the presence of rearrangements, particularly in Sox17-3 subclone #1 and Sox17-1 pool. For BAC5 clones, fragments of the BAC were present at single copy, suggesting that those sequences were deleted on those E-BACs (Figure 3B). Most likely, these genetic alternations occurred in E. coli and were present at low abundance in the pool, but we cannot rule out that they could have been established post-transfection independently in each clone. Regardless, clones exhibit some genetic variation from each other and from the pool, which must be taken into account in interpreting results. These results confirm that E-BACs can be maintained as extra-chromosomal circular units in HeLa-EBNA1 cells.
Figure 3.
E-BACs are maintained as extra-chromosomal units in Hela-EBNA1 cells at different copy numbers. (A) Gardella gel (Sox17-3 and sox17-1) or Hirt extraction (BAC5) results for E-BAC transfected HeLa-EBNA1 cells (Gardella and Hirt extraction give similar results, Supplementary Figure S3A). The circular E-BAC control (E-BAC Circular) was prepared by lysing sox17-1 carrying E. coli embedded in an agarose block; linear E-BAC control (E-BAC linear) is Mlu1-linearized sox17-1 E-BAC, and DNA from E-BAC-free HeLa-EBNA1 (HeLa-EBNA1) served as the negative control. DNA was hybridized with a 5kb probe derived from BAC vector pBACe3.6. (B) Total Repli-seq read coverage (for early + late S fractions, Figure 4A) for E-BAC regions. The numbers on each plot represent copy number estimation of each E-BAC (see Materials and Methods). Blue arrows indicate deletions in the E-BAC5 clones.
E-BAC replication timing
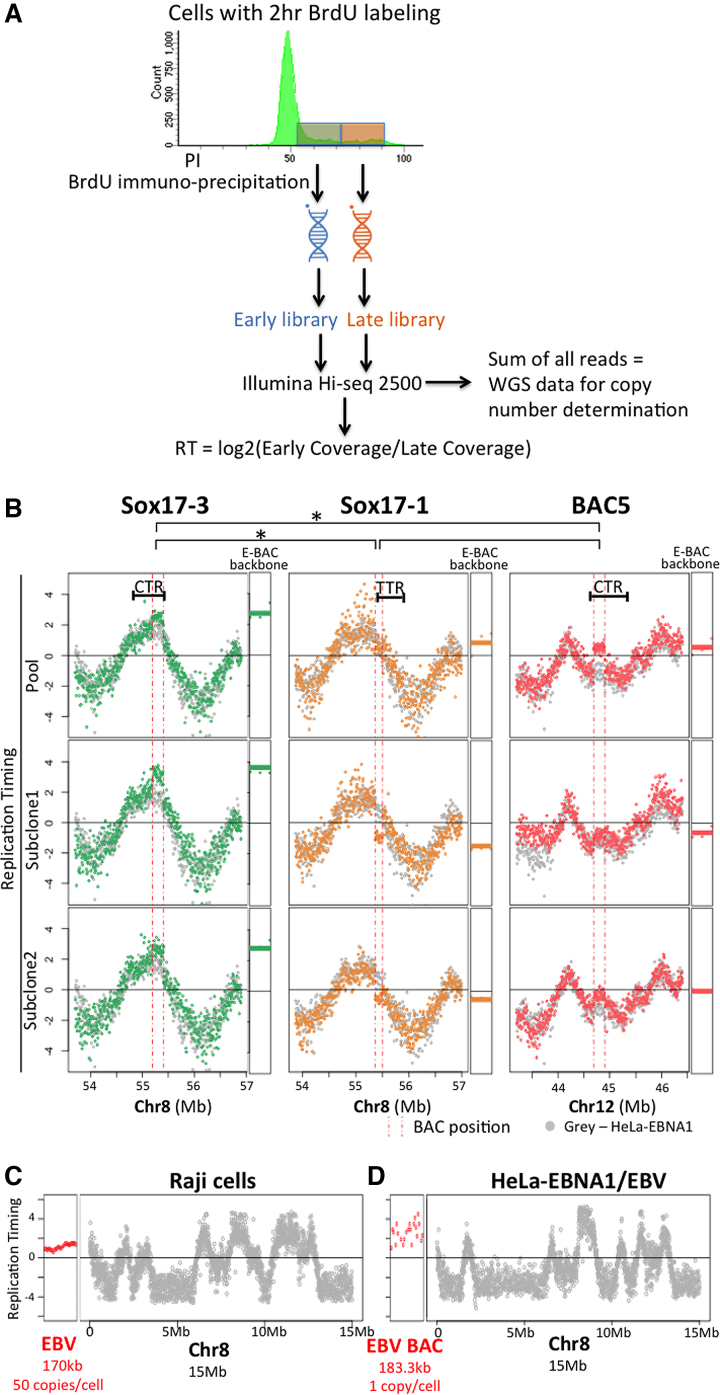
We performed repli-seq to assay RT of the E-BACs (Figure 4A). Briefly, cells were pulse labeled with BrdU for 2 h, and FACS (Fluorescence Activated Cell Sorting) sorted into early vs late S phase by DNA content. BrdU substituted DNA was then immunoprecipitated, amplified, and sequenced with ∼10 million mappable reads per fraction (Figure 4A). We plotted the log2 ratios of early versus late coverage for 6 kb non-overlapping bins. Results revealed that the Sox17-3 E-BAC (derived from an early replicating CTR) replicated early in S phase in both the pool and the two clones, including Subclone #1 that showed a high degree of variation in read coverage (Figure 4B). However, the Sox17-1 BAC replicated in mid-late S phase, despite sharing 45.9 kb of sequence with Sox17-3. Sox17-1 BAC sequences are derived from a TTR, which is normally replicated early from forks emanating from the CTR corresponding to Sox17-3 BAC sequences. These results suggest that the early replicating determinants in Sox17-3 BAC are not contained within these 45.9 kb. The Sox17-1 BAC pool replicated slightly earlier than the two clones, which may be related to its higher variability in read coverage, since both clones showed similar and uniform read coverage indicating that they contain largely intact BAC DNA. BAC5 (derived from a late replicating CTR) also replicated in mid-late S phase (Figure 4B).
Figure 4.
E-BAC replication timing. (A) Experimental flowchart for Repli-seq. Exponentially growing cells were labeled with BrdU for 2 h, sorted for DNA content into early versus late S phase. DNA was isolated, sheared and the nascent DNA synthesized either early or late during S phase was immunoprecipitated with an anti-BrdU antibody. BrdU-substituted DNA was sequenced and the log2 ratio of sequences in early versus late S phase fractions was calculated in 6 kb non-overlapping windows. To determine copy number, the reads from early and late fractions were summed to acquire whole genome sequencing information and the total number of reads per 6kb window was determined (see Figure 3B and Materials and Methods). (B) E-BAC repli-seq profiles. The Y-axis for each data point is the log2 ratio for reads within a 6 kb window. HeLa-EBNA1 cells without E-BAC transfection were used as control (gray data points). Vertical pink dashed lines indicate the human chromosomal map positions of the segments contained in each BAC. RT of the 21 kb bacterial sequences of each E-BAC (pBACe3.6 E-BAC backbone) was plotted separately and indicated as horizontal lines (average of all 6kb windows) next to the chromosomal regions. (C, D) Replication timing of the EBV genome in Raji cells (C) and the EBV BAC in HeLa-EBNA1/EBV (D) (red data points). The datapoints for HeLa-EBNA1 EBV BAC are more dispersed because there are fewer total reads due to the substantially lower copy number of the EBV sequences in this cell line (Supplementary Figure S9). Data from 15 Mb of chr8 is plotted for comparison (grey data points).
To quantify the significance of differences in RT, we performed paired, one-tailed Student's t-test on RT mean values of the E-BACs and E-BAC backbone from pool and clones of the three E-BACs. These results demonstrated that RT of sox17-3 E-BAC is significantly different than sox17-1 (P = 0.00062) and BAC5 (P = 0.00024), whereas RT of sox17-1 and BAC5 are not statistically different (P = 0.11). However, these RT values are not quantitatively the same as the endogenous locus. Statistically significant differences were found between Sox17-3 (E-BAC earlier; P = 0.0063) and Sox17-1 (E-BAC later; P = 0.0094) but not E-BAC5 (P = 0.41) and their corresponding chromosomal loci. As a control, we also performed repli-seq analysis of Raji cells (48), an EBV transformed human lymphoid cell line that carries more than 50 copies of the latent virus, as well as HeLa-EBNA1 cells transfected with a BAC containing the entire EBV genome (49) (gift from Wolfgang Hammerschmidt lab), and found that EBV replicates in early-middle S phase in both of these contexts (Figure 4C, D and Supplementary Figure S9). These control experiments suggest that RT of our E-BACs is not dictated by the presence of EBV FR sequences. Altogether, these results provide evidence that the ∼200 kb DNA segments contained within each BAC are sufficient to establish RT patterns de novo that are qualitatively consistent with endogenous loci and different from the RT of native EBV. The E-BAC derived from an early CTR replicates early while the E-BACs from a TTR or a late CTR replicate with a mid-late pattern and with more RT variability.
E-BAC compartment analysis
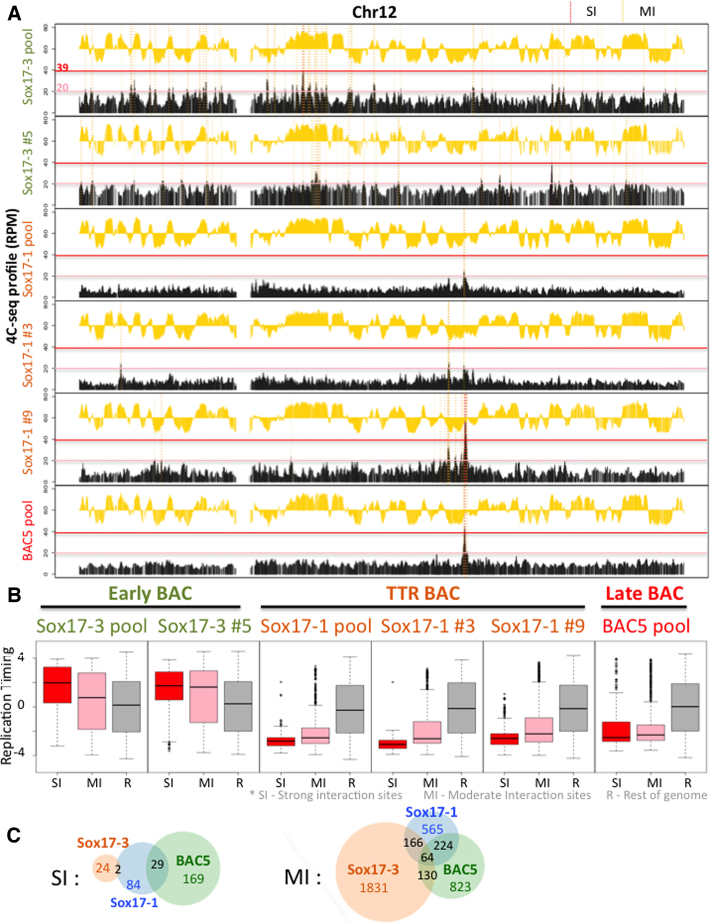
To determine whether E-BACs become compartmentalized in the nucleus, we performed 4C-seq (42) using a bait on the pBAC3.6 plasmid backbone in common to each BAC. Three to seven million mappable reads were obtained for each profile. Since the presence of integrated BACs is easily detected as an extremely high interaction frequency with sequences flanking the ectopic integration site, we first scanned the genome for such high frequency interaction profiles. Results confirmed that most of the E-BACs remain extra-chromosomal, as the strongly enriched reads were restricted to the genomic sequences contained within each E-BAC. One exception was the BAC5 pool, which had a clear signature of integration at a specific site on chr14 (Supplementary Figure S4). From browsing 4C-seq profiles, the early replicating E-BAC 4C-seq profiles have a broad distribution of interaction sites across the genome, whereas the TTR and late replicating E-BACs interact strongly with several specific sites (Figure 5A and Supplementary Figure S6). Next, we divided the 4C-seq interaction peaks into two thresholds: strong interaction sites (SIs) and moderate interaction sites (MIs). SIs were defined as peaks with interaction scores from 39 to 400 (RPM, 400 ceiling was chosen to remove the very strong interactions near the integration site for BAC5 E-BAC), which could be frequent interaction sites or could also represent integration sites that are present in a small percentage of cells. MIs are peaks with interaction scores between 39 and 20 (Supplementary Figure S6). Both SIs and MIs of the early sox17-3 BAC were strongly enriched in early replicating regions, while the sox17-1 and BAC5 E-BACs have both types of sites enriched in the late compartment (Figure 5). This trend is preserved in both the pool and the clones (Figure 5) and was similar using different cutoffs for defining SIs and MIs (Supplementary Figure S6D). Interestingly, sox17-1 and BAC5 E-BACs shared a large percentage of specific interaction sites (Figure 5C). This overlap was highly significant as measured by Jaccard similarity index, which was 12.9% for SIs and 19.2% for MIs between Sox17-1 and BAC5, in comparison to 0–5.2% between sox17-3 and BAC5 and 1.9% to 7.4% between sox17-3 and sox17-1. These sites are not specific to EBNA1 binding sites in the genome (EBNA1 ChIP in Raji cells) (51), and it will be interesting to explore the mechanistic underpinnings of these common interaction sites. Altogether, 4C analysis demonstrates that E-BACs establish de novo interactions with chromatin in the sub-nuclear compartment that is appropriate for their respective replication times.
Figure 5.
E-BAC compartment analysis. 4C-seq was performed using a bait on the pBACe3.6 plasmid backbone. (A) Exemplary 4C-seq and repli-seq profiles for chr12 are plotted as a black or yellow histogram, respectively. Strong Interaction sites (SIs) with reads between 39 to 400 RPM, and Moderate Interaction site (MIs) with reads between 20 and 39 RPM, are shaded by red or yellow dashed vertical lines, respectively, demonstrating how the strong and moderate interaction sites were defined for B and C. The red and pink horizontal lines indicate the y-axis positions of the SI and MI cut-offs. (B) Boxplots demonstrate RT distribution of E-BAC Strong Interaction sites (SI, red box), Moderate Interaction sites (MI, pink box) and the rest of genome as control (R, gray box) for all three pools and Sox17-3 subclone 5 (Sox17-3 #5), Sox17-1 clones 3 and 9 (Sox17-1 #3 and #9). (C) Venn diagram shows the number of shared SI or MI sites (50kb windows) in 4C-seq profiles of Sox17-3, Sox17-1 and BAC5 pool.
A late E-BAC maintains late replication in human induced pluripotent cells
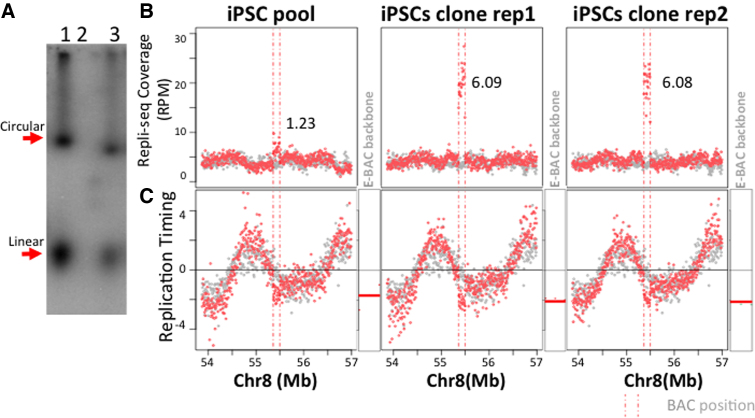
To test whether the E-BAC system works in other cell types, we made stable EBNA1 expressing human induced pluripotent cells (hiPSCs), using hiPSC line K3 (Supplementary Figure S7), and introduced E-BAC sox17-1. Because the TK promoter, driving hygR in our E-BACs, is a weak promoter in pluripotent stem cells, we replaced the TK-hygR gene in the FR cassette (Figure 1) with CAG-GFP and FACS enriched GFP positive cells instead of drug selection. Gardella gel (Figure 6A) and 4C-seq results (Supplementary Figure S8A) confirmed that the sox17-1 E-BAC is maintained extrachromosmally. Since we found that the pool had only 1–2 copies of the E-BAC, we picked a GFP-positive colony, which was then expanded, BrdU-labeled, and sorted additionally for bright GFP-expressing cells by FACS immediately prior to repli-seq. This yielded a population of cells with 6 copies per cell on average (Figure 6B). Both pool and clones demonstrated unaltered pluripotency based on their genome-wide RT profiles, which are a strong cell type specific marker of pluripotency as they change rapidly upon differentiation (Supplementary Figure S8B and C). Repli-seq results revealed that sox17-1 E-BAC maintained late replication in both the pool and the subclone, consistent with that of the endogenous locus (Figure 6C). These results suggest that RT can be maintained on extrachromosomal vectors in pluripotent cells and the E-BAC system can be used to study chromosome structure and function in stem cells.
Figure 6.
Replication timing of sox17-1 E-BAC in K3-EBNA1 hiPSCs. (A) Gardella gel result for Sox17-1 E-BAC transfected K3-EBNA1 pool. Lane 1: Sox17-1 E-BAC transfected HeLa-EBNA1 cells; Lane 2: K3-EBNA1 cells; Lane 3: Sox17-1 E-BAC transfected K3-EBNA1 pool. (B) Repli-seq read coverage for E-BAC regions. The numbers on each plot represent copy number estimation of Sox17-1 E-BAC (see Materials and Methods). K3-EBNA1 without E-BAC transfection was used as control (grey data points). (C) E-BAC replication timing in K3-EBNA1 cells. Log2 ratio of early to late coverage of K3-EBNA1 with sox17-1 E-BAC was plotted (red data points) as compared to K3-EBNA1 cells as control (gray data points). Vertical pink dashed lines indicate the Sox17-1 BAC corresponding position on chr8. RT of the 21 kb bacterial sequences of each E-BAC (pBAC3.6 E-BAC backbone) was plotted separately and indicated as horizontal lines next to the chromosomal regions (as in Figure 4B).
DISCUSSION
We have developed an extra-chromosomal vector system that enables functional dissection of genomic DNA. We have shown that E-BACs can be stably maintained as extra-chromosomal replicating units in EBNA1-expressing cells and the 200 kb of DNA they harbor is sufficient to dictate their RT and genome compartmentalization. An E-BAC derived from an early replicating region maintains early replication, and preferentially interacts with early replicating regions. In contrast, E-BACs corresponding to a TTR or a late CTR replicate in mid- to late-S phase and interact with the late replicating chromatin compartment, consistent with our previous finding that TTRs share chromatin properties with late regions (52,53). A previous report has shown that double minute chromosomes replicate at a similar time as their genomic counterparts (54), but this study could not distinguish whether RT was transferred epigenetically from the original chromatin or derived from the underlying DNA sequences. Our results suggest that ∼200 kb DNA sequences can harbor sufficient information to direct appropriate RT and compartmentalization. Altogether, we describe a versatile system derived from EBV with which BACs of interests can be stably maintained as autonomous replicating units in EBNA1 expressing human cells including pluripotent stem cells.
The RT of full length EBV has been somewhat controversial, with studies reporting either early (55–57) or late replication (54,58), even between different groups working with Raji cells. Our results find that EBV replicates during early-mid S phase in Raji cells, as well as HeLa EBNA1. We do not know the source of the discrepancy, but we find that genome-wide analyses, where the RT of each genomic window validates the RT of the adjacent genomic window, are more reproducible and reliable than the single site Q-PCR assays done in the past. Moreover, our results in both HeLa EBNA1 and Raji cells are consistent with a previous report of latent EBV’s association with regions of decondensed, presumably early replicating, chromatin in primary human B lymphocytes (50). This early-mid replication is likely contributed by EBV genomic sequences other than the FR carried on our vectors, as small FR plasmids replicate in late S phase in 293 cells (34). In any case, since our three E-BACs replicate at different times, one of which is significantly earlier than the replication time of full length EBV viral genomes, it is unlikely that E-BAC replication timing and genome compartment are dictated by the supplied EBV viral retention system (EBNA1 + FR). Moreover, our 4C-seq results with different E-BACs show differential compartmentalization of early vs. late replicating E-BACs, suggesting that the genomic DNA segments carried on E-BAC vectors can override both the natural RT of EBV and any chromatin interaction preferences. Presumably, cis-elements within those genomic segments control the RT and compartmentalization of the E-BACs.
Transcription has long been correlated with early replication, but causal relationships between the two have not been established (10). All of our E-BAC transfected HeLa-EBNA1 cells were grown under hygromycin B selection requiring that the hygR gene be expressed. However, the E-BACs manifested distinct RT and compartment patterns. Similarly, the sox17-1 E-BAC steadily expresses GFP in hiPSCs, yet it was clearly late replicating. Therefore, our results support the conclusion from many other correlative studies that gene transcription is not sufficient to dictate early replication or sub-nuclear position (59).
Our results also raise several interesting questions. First, what are the sequence features underlying early versus TTR/late sequences that give rise to higher order properties such as RT and sub-nuclear localization? Second, how do those higher properties relate to other 2D chromatin features such as nucleosome occupancy and histone modifications? It is as yet unknown whether the same chromatin marks are deposited on E-BACs as compared to their genomic counterparts, and if those marks are cause or effect of compartmentalization. Third, previous studies have characterized sequences from the beta-globin locus that are sufficient for dictating early replication of an ectopic chromosomal insertion sites in avian and mammalian cells (60,61). Whether such discrete functional elements exist on each BAC and what their role may be in replication remain to be examined. Our results demonstrate that E-BACs show promise to address these questions by providing a relatively facile method to study large-scale chromosome structure and function.
AVAILABILITY
The HeLa repli-seq and 4C-seq data from this study have been submitted to the database of Genotypes and Phenotypes (dbGaP), as a substudy under accession number phs000640. The K3 repli-seq and 4C-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSE102704.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Bill Sudgen, Wolfgang Hammerschmidt, Michele P. Calos and Aloys Schepers for sharing plasmids/BACs generously and gave constructive advices on the project. We thank Paul M. Lieberman for sharing Raji cells, Vishnu Dileep for suggestions on 4C-seq data analysis, Carley Huffstetler for modifying the figures, Ruth Dider for florescent-activated cell sorting, Peiyao Zhao, Ariana Belsky and Robinson Blake for helping with BAC isolation, Rajkumari Gayatri for help with Gardella gels, Denis Avery, Brian Washburn, Cheryl Pye and Kristina Poduch for vector cloning, Fanxiu Zhu for helpful discussions.
The genome sequence described/used in this research was derived from a HeLa cell line. Henrietta Lacks, and the HeLa cell line that was established from her tumor cells without her knowledge or consent in 1951, have made significant contributions to scientific progress and advances in human health. We are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research. The data generated from this research were submitted to the database of Genotypes and Phenotypes (dbGaP), as a substudy under accession number phs000640.
Authors Contributions: J.S. conducted the majority of the experiments and analyzed the data. D.B. performed the 4C-seq experiments. M.G. performed the experiments in hiPSCs. J.S. and D.M.G. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM083337, GM085354 to D.M.G.]. Funding for open access charge: NIH [GM083337, GM085354 to D.M.G.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rhind N., Gilbert D.M.. DNA replication timing. Cold Spring Harb. Perspect. Med. 2013; 3:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hiratani I., Ryba T., Itoh M., Rathjen J., Kulik M., Papp B., Fussner E., Bazett-Jones D.P., Plath K., Dalton S. et al. . Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010; 20:155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J. et al. . Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012; 485:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K. et al. . Topologically associating domains are stable units of replication-timing regulation. Nature. 2014; 515:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ryba T., Hiratani I., Lu J., Itoh M., Kulik M., Zhang J., Schulz T.C., Robins A.J., Dalton S., Gilbert D.M.. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010; 20:761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W. et al. . Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008; 453:948–951. [DOI] [PubMed] [Google Scholar]
- 8. Wang S., Su J.-H., Beliveau B.J., Bintu B., Moffitt J.R., Wu C., Zhuang X.. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016; 353:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Solovei I., Thanisch K., Feodorova Y.. How to rule the nucleus: divide et impera. Curr. Opin. Cell Biol. 2016; 40:47–59. [DOI] [PubMed] [Google Scholar]
- 10. Rivera-Mulia J.C., Gilbert D.M.. Replication timing and transcriptional control: beyond cause and effect—part III. Curr. Opin. Cell Biol. 2016; 40:168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sima J., Gilbert D.M.. Complex correlations: replication timing and mutational landscapes during cancer and genome evolution. Curr. Opin. Genet. Dev. 2014; 25:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Supek F., Lehner B.. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015; 521:81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ryba T., Battaglia D., Chang B.H., Shirley J.W., Buckley Q., Pope B.D., Devidas M., Druker B.J., Gilbert D.M.. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome Res. 2012; 22:1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sasaki T., Rivera-Mulia J.C., Vera D., Zimmerman J., Das S., Padget M., Nakamichi N., Chang B.H., Tyner J., Druker B.J. et al. . Stability of patient-specific features of altered DNA replication timing in xenografts of primary human acute lymphoblastic leukemia. Exp. Hematol. 2017; 51:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pope B.D., Chandra T., Buckley Q., Hoare M., Ryba T., Wiseman F.K., Kuta A., Wilson M.D., Odom D.T., Gilbert D.M.. Replication-timing boundaries facilitate cell-type and species-specific regulation of a rearranged human chromosome in mouse. Hum. Mol. Genet. 2012; 21:4162–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sinclair P., Bian Q., Plutz M., Heard E., Belmont A.S.. Dynamic plasticity of large-scale chromatin structure revealed by self-assembly of engineered chromosome regions. J. Cell Biol. 2010; 190:761–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Erliandri I., Fu H., Nakano M., Kim J.-H., Miga K.H., Liskovykh M., Earnshaw W.C., Masumoto H., Kouprina N., Aladjem M.I. et al. . Replication of alpha-satellite DNA arrays in endogenous human centromeric regions and in human artificial chromosome. Nucleic Acids Res. 2014; 42:11502–11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van de Werken H.J.G., Haan J.C., Feodorova Y., Bijos D., Weuts A., Theunis K., Holwerda S.J.B., Meuleman W., Pagie L., Thanisch K. et al. . Small chromosomal regions position themselves autonomously according to their chromatin class. Genome Res. 2017; 27:922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adams A., Lindahl T.. Epstein-Barr virus genomes with properties of circular DNA molecules in carrier cells. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:1477–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hammerschmidt W., Sugden B.. Replication of Epstein-Barr Viral DNA. Cold Spring Harb. Perspect. Biol. 2013; 5:a013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yates J., Warren N., Reisman D., Sugden B.. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:3806–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yates J.L., Warren N., Sugden B.. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 313:812–825. [DOI] [PubMed] [Google Scholar]
- 23. Reisman D., Yates J., Sugden B.. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 1985; 5:1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frappier L., O’Donnell M.. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:10875–10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frappier L. EBNA1 and host factors in Epstein-Barr virus latent DNA replication. Curr. Opin. Virol. 2012; 2:727–733. [DOI] [PubMed] [Google Scholar]
- 26. Kanda T., Kamiya M., Maruo S., Iwakiri D., Takada K.. Symmetrical localization of extrachromosomally replicating viral genomes on sister chromatids. J. Cell Sci. 2007; 120:1529–1539. [DOI] [PubMed] [Google Scholar]
- 27. Norio P., Schildkraut C.L.. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science. 2001; 294:2361–2364. [DOI] [PubMed] [Google Scholar]
- 28. Norio P., Schildkraut C.L.. Plasticity of DNA replication initiation in Epstein-Barr virus episomes. PLoS Biol. 2004; 2:816–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Little R.D., Schildkraut C.L.. Initiation of latent DNA replication in the epstein-barr virus genome can occur at sites other than the genetically defined origin. Mol. Cell. Biol. 1995; 15:2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Papior P., Arteaga-Salas J.M., Günther T., Grundhoff A., Schepers A.. Open chromatin structures regulate the efficiencies of pre-RC formation and replication initiation in Epstein-Barr virus. J. Cell Biol. 2012; 198:509–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Norio P., Schildkraut C.L., Yates J.L.. Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein-Barr virus chromosome after infection of established cell lines. J. Virol. 2000; 74:8563–8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ott E., Norio P., Ritzi M., Schildkraut C., Schepers A.. The dyad symmetry element of Epstein-Barr virus is a dominant but dispensable replication origin. PLoS One. 2011; 6:e18609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heinzel S.S., Krysan P.J., Tran C.T., Calos M.P.. Autonomous DNA replication in human cells is affected by the size and the source of the DNA. Mol. Cell. Biol. 1991; 11:2263–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haase S.B., Calos M.P.. Replication control of autonomously replicating human sequences. Nucleic Acids Res. 1991; 19:5053–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krysan P.J., Calos M.P.. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol. Cell. Biol. 1991; 11:1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wade-Martins R., White R.E., Kimura H., Cook P.R., James M.R.. Stable correction of a genetic deficiency in human cells by an episome carrying a 115 kb genomic transgene. Nat. Biotechnol. 2000; 18:1311–1314. [DOI] [PubMed] [Google Scholar]
- 37. Black J., Vos J.-M.. Establishment of an oriP/EBNA1-based episomal vector transcribing human genomic beta-globin in cultured murine fibroblasts. Gene Ther. 2002; 9:1447–1454. [DOI] [PubMed] [Google Scholar]
- 38. Sun T.Q., Fenstermacher D.A., Vos J.M.H.. Human artificial episomal chromosomes for cloning large DNA fragments in human cells. Nat. Genet. 1994; 8:33–41. [DOI] [PubMed] [Google Scholar]
- 39. Westphal E., Sierakowska H.. A system for shuttling 200 kb BAC/PAC clones into human cells: stable extrachromosomal persistence and long-term ectopic gene activation. Hum. Gene Ther. 1998; 1873:1863–1873. [DOI] [PubMed] [Google Scholar]
- 40. Gardella T., Medveczky P., Sairenji T., Mulder’ C.. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J. Virol. 1984; 50:248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adey A., Burton J.N., Kitzman J.O., Hiatt J.B., Lewis A.P., Martin B.K., Qiu R., Lee C., Shendure J.. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013; 500:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Splinter E., de Wit E., van de Werken H.J.G., Klous P., de Laat W.. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods. 2012; 58:221–230. [DOI] [PubMed] [Google Scholar]
- 43. Griffin B.E., Bjorck E., Bjursell G., Lindahl T.. Sequence complexity of circular Epstein-Barr virus DNA in transformed cells. J.Virol. 1981; 40:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hiratani I., Ryba T., Itoh M., Rathjen J., Kulik M., Papp B., Fussner E., Bazett-Jones D.P., Plath K., Dalton S. et al. . Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010; 20:155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dileep V., Rivera-Mulia J.C., Sima J., Gilbert D.M.. Large-scale chromatin structure-function relationships during the cell cycle and development: insights from replication timing. Cold Spring Harb. Symp. Quant. Biol. 2015; 80:53–63. [DOI] [PubMed] [Google Scholar]
- 46. Bian Q., Khanna N., Alvikas J., Belmont A.S.. β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J. Cell Biol. 2013; 203:767–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meuleman W., Peric-Hupkes D., Kind J., Beaudry J.-B., Pagie L., Kellis M., Reinders M., Wessels L., van Steensel B.. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013; 23:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pulvertaft R.J.V. Cytology of Burkitt's tumour (African lymphoma). Lancet. 1964; 283:238–240. [DOI] [PubMed] [Google Scholar]
- 49. Delecluse H.J., Hilsendegen T., Pich D., Zeidler R., Hammerschmidt W.. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deutsch M.J., Ott E., Papior P., Schepers A.. The latent origin of replication of Epstein-Barr virus directs viral genomes to active regions of the nucleus. J. Virol. 2010; 84:2533–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tempera I., De Leo A., Kossenkov A. V., Cesaroni M., Song H., Dawany N., Showe L., Lu F., Wikramasinghe P., Lieberman P.M.. Identification of MEF2B, EBF1, and IL6R as direct gene targets of Epstein-Barr Virus (EBV) nuclear antigen 1 critical for EBV-infected B-lymphocyte survival. J. Virol. 2016; 90:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K. et al. . Topologically associating domains are stable units of replication-timing regulation. Nature. 2014; 515:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dileep V., Ay F., Sima J., Vera D.L., Noble W.S., Gilbert D.M.. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res. 2015; 25:1104–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carroll S.M., Trotter J., Wahl G.M.. Replication timing control can be maintained in extrachromosomally amplified genes. Mol. Cell. Biol. 1991; 11:4779–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hampar B., Tanaka A., Nonoyama M., Derge J.G.. Replication of the resident repressed Epstein-Barr virus genome during the early S phase (S-1 period) of nonproducer Raji cells. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:631–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Benz W.C., Strominger J.L.. Viral and cellular DNA synthesis in nuclei from human lymphocytes transformed by Epstein-Barr virus. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:2413–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vogel B., Full F., Biesinger B., Linden C., Alberter B., Ensser A.. Episomal replication timing of γ-herpesviruses in latently infected cells. Virology. 2010; 400:207–214. [DOI] [PubMed] [Google Scholar]
- 58. Zhou J., Snyder A.R., Lieberman P.M.. Epstein-Barr virus episome stability is coupled to a delay in replication timing. J. Virol. 2009; 83:2154–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Harr J.C., Luperchio T.R., Wong X., Cohen E., Wheelan S.J., Reddy K.L.. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol. 2015; 208:33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hassan-Zadeh V., Chilaka S., Cadoret J.-C., Ma M.K.-W., Boggetto N., West A.G., Prioleau M.-N.. USF binding sequences from the HS4 insulator element impose early replication timing on a vertebrate replicator. PLoS Biol. 2012; 10:e1001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Aladjem M.I., Rodewald L.W., Kolman J.L., Wahl G.M.. Genetic dissection of a mammalian replicator in the human β-globin locus. Science. 1998; 281:1005–1009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.