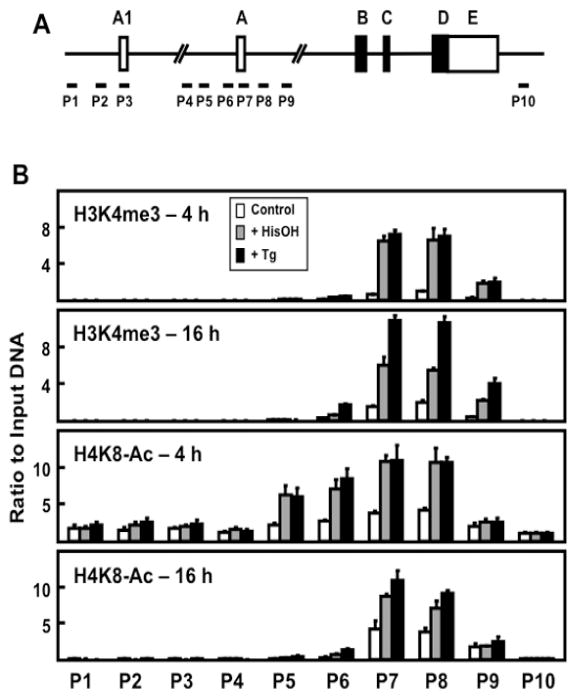
Figure 6.
Analysis of H3K4me3 and H4K8Ac modifications across the ATF3 gene locus during the AAR and UPR pathways. (A) Schematic showing ATF3 gene structure and primer locations for ChIP analysis at specific regions of the gene locus. (B) HepG2 cells were cultured in DMEM ± 2.5 mM HisOH or ± 50 nM Tg for 4 or 16 h. The histone modifications H3K4me3 and H4K8-Ac were analyzed by ChIP at the ATF3 gene regions indicated. The data are plotted as the ratio to input DNA and the values for histone modifications were normalized to the signal obtained for total histone H3 at the respective regions (Figure 5). Values are means ± S.D. for at least three replicates and each experiment was repeated to ensure reproducibility between batches of cells.