Fig. 1. AMPK regulated a nucleosome remodeling network by phosphorylating DNMT1, RBBP7, and HAT1.
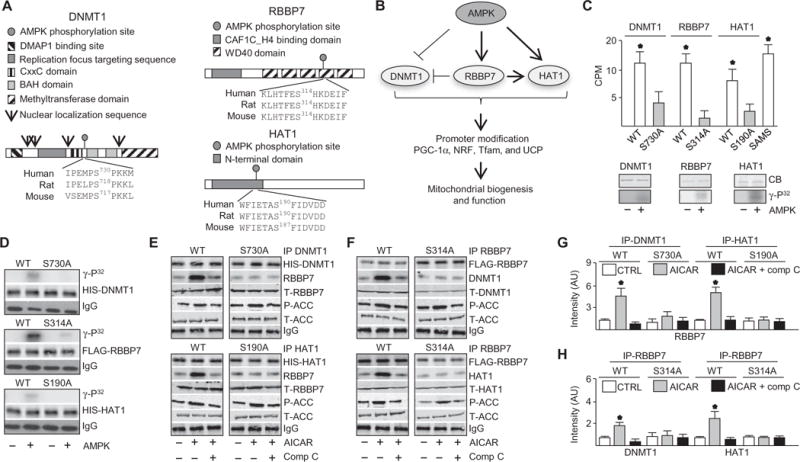
(A) Protein domains and putative AMPK-mediated phosphorylation sequence of DNMT1, RBBP7, and HAT1. DMAP1, DNA methyltransferase 1–associated protein 1 binding domain; BAH, bromo-adjacent homology domain. (B) Illustrated hypothesis of AMPK-mediated phosphorylation of DNMT1, RBBP7, and HAT1 and effects on mitochondrial biogenesis and function. (C) Kinase assays using recombinant target protein in the presence or absence of activated recombinant AMPKα2β1γ1. Top: Kinase assays conducted with peptides. Bottom: Kinase assays with full-length proteins. n = 4 independent experiments. CPM, counts per minute. (D) Kinase assays using immunoprecipitated wild-type (WT) and mutated DNMT1, RBBP7, and HAT1 proteins from HUVECs. Top: Autoradiograph. Lower: Total protein and immunoglobulin G (IgG) immunoblots for loading. n = 3 independent experiments. (E and F) Coimmunoprecipitation (IP) immunoblots in HUVECs transfected with WT DNMT1, RBBP7, or HAT1 or their corresponding Ser-to-Ala mutants (DNMT1-S730A, HAT1-S190A, and RBBP7-S314A) and treated with AICAR or left untreated for 30 min (top) or 10 min (bottom). P-ACC (phospho–acetyl-CoA carboxylase), T-ACC (total acetyl-CoA carboxylase), T-RBBP7, T-DNMT1, and T-HAT1 immunoblotting was conducted with the input IP crude cell lysate. n = 4 independent experiments. Comp C, compound C. (G and H) Densitometry analysis of coimmunoprecipitation immunuoblots comparing coimmunoprecipitated protein to total protein. *P < 0.05. AU, arbitrary units.
