Figure 3.
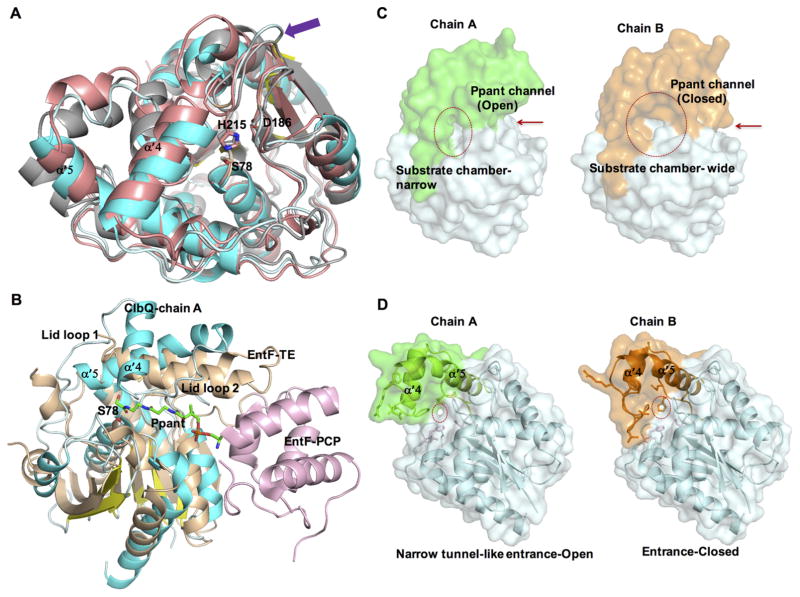
Conformational flexibility of ClbQ: (A) Alignment of ClbQ (chain A, cyan) with RedJ (salmon, PDB code 3QMV, chain B) and RifR (gray, PDB code 3FLB, chain A). The extended loop region in ClbQ formed by the motif PAADH is indicated (purple arrow). The catalytic triad is labeled. (B) Modeling of a Ppant arm (green) into ClbQ active site based on the EntF PCP (light pink)–TE (wheat) didomain structure (PDB code 3TEJ) with ClbQ (chain A, blue/cyan). (C) Surface diagram of the two monomers of ClbQ. The substrate binding site is indicated by a red circle, and Ppant entrance is indicated by red arrow. (D) Cartoon and surface representative of the active site entrance channel. The narrow tunnel-like entrance channel of chain A (circled and open) and chain B (closed, largely by His115 and Glu116).