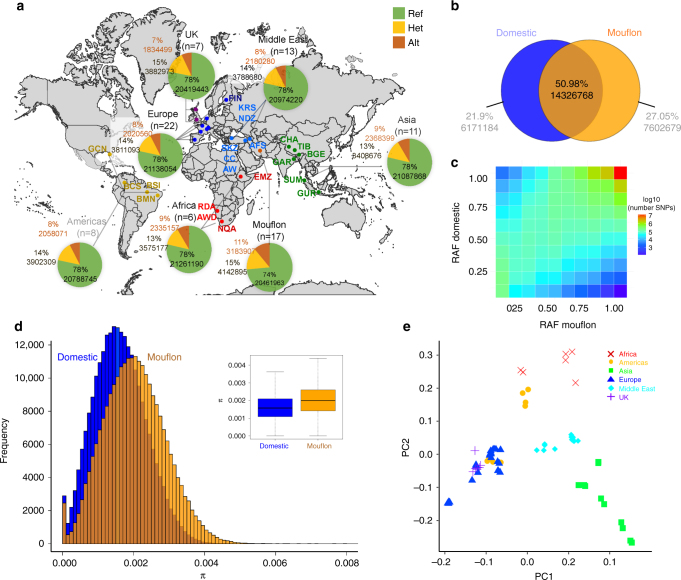
Fig. 1.
Genome diversity and relatedness of wild and domestic sheep. a The geographic distribution of 43 breeds sampled for whole-genome sequencing. Population based proportion of allele type is given for each major geographic group of breeds, after calling reference variants present in the Texel derived genome assembly OARv3.1. b Proportion and number of private and shared SNP for the collection of wild and domestic sheep genomes. c Reference allele frequency (RAF) correlation between domestic and mouflon populations. RAF domestic and mouflon represent the frequency of the reference allele in domestic and mouflon populations respectively. (Pearson correlation: 76.36% p-value <2.2E−16). d Distribution of nucleotide diversity (π) within species, estimated within 20 kb bins. The boxplot compares the bin mean and variance. The distribution means, evaluated using the Wilcoxon rank-sum test, are significantly difference (p-value <2.2E−16). e Principal component (PC) analysis clustering individual domestic sheep coloured to reflect the geographic origin of breed development