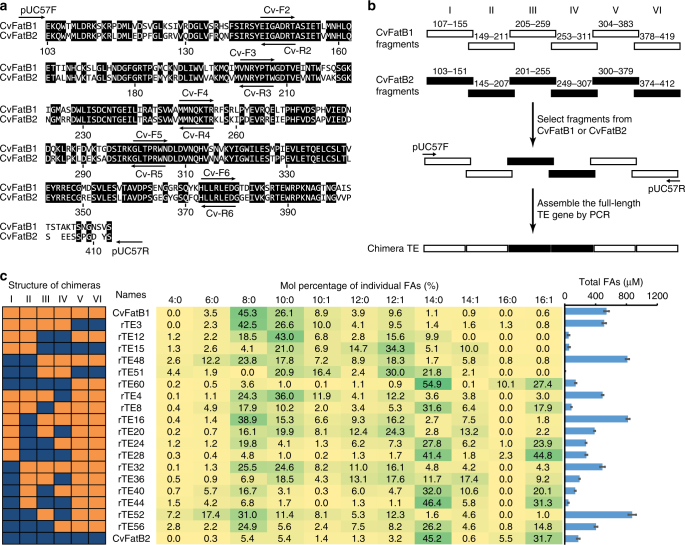
Fig. 1.
Domain-swapping to identify segments that determine substrate specificity. a Sequence alignment of CvFatB1 and CvFatB2 identifying identical residues shared between the two enzymes, and the positions of DNA primers used to construct chimeric enzymes; b schematic diagram illustrating the construction of chimeric acyl-ACP TEs; c substrate specificities of wild-type and chimeric acyl-ACP TEs. The sequence structures of the chimeric acyl-ACP TEs are presented as different color combinations; orange represents the CvFatB1-sourced fragment, and blue represents the CvFatB2-sourced fragment. The central table shows the fatty acid profile generated by the E. coli strains expressing each chimeric enzyme, and green shading highlights the major fatty acids produced by each chimera. The graph on the right identifies the total free FAs accumulated in the medium. The data are the average of four replicates, and the error bar represents standard error of the mean