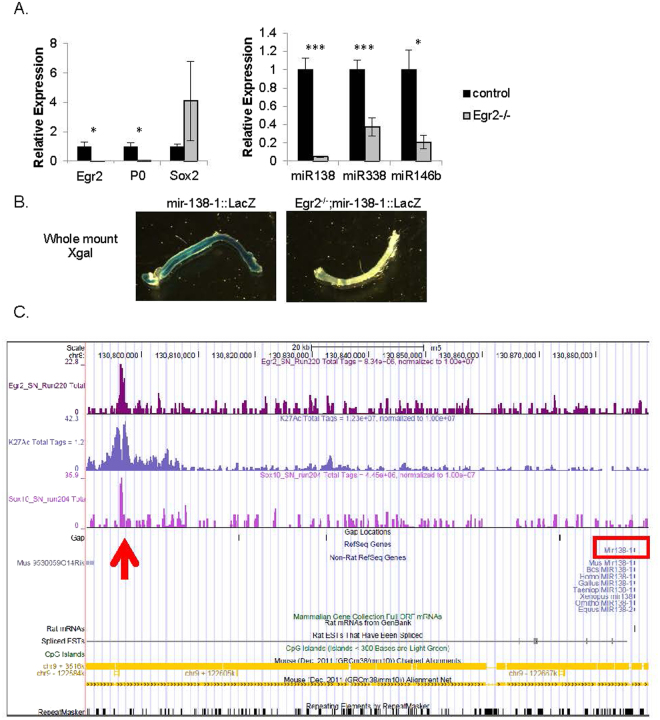
Figure 3.
Egr2 is required for mir-138-1 transcription during development and both SOX10 and EGR2 bind to an active enhancer near the mir-138-1 locus. (A) Quantitative RT-PCR expression levels of selected SC genes (Mpz, Egr2 and Sox2) and microRNA candidates (miR-138, 338-3p and 146b) in Egr2-null sciatic nerves (gray bars) and littermate controls (black bars) at P5 (*p < 0.05, **p < 0.001, ***p < 0.005). (B) Shown are whole mount Xgal staining of P7 mir-138-1::LacZ and Egr2−/−; mir-138-1::LacZ sciatic nerves. Blue Xgal staining reports endogenous transcription activities at mir-138-1 locus. Transcription activity at mir-138-1 at P7 is detectable in control sciatic nerve but is not detectable in Egr2−/− sciatic nerves. (C) The UCSC genome browser view shown in the figure displays peaks of SOX10 and EGR2 binding based on analysis of the ChIP-Seq data from pooled P15 rat sciatic nerves25. Histone H3 K27Ac data is a track of active enhancers also obtained from P15 sciatic nerves. The peaks are co-localized (red arrow) and within 100 kb from mir-138-1 locus (red box).