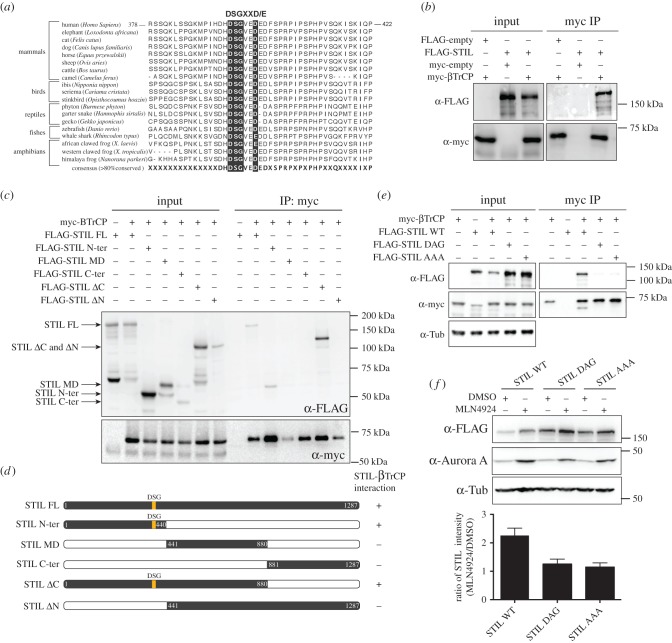
Figure 2.
(a) Multiple protein sequence alignment (ClustalW) of vertebrate STIL sequences. The consensus sequence (more than 80% conserved) is shown at the bottom and taxonomic classes are shown on the side. The region encompassing the DSG motif is highlighted with black boxes. (b) HEK 293T cells overexpressing FLAG-STIL and myc-βTrCP for 36 h were lysed and subjected to immunoprecipitation (IP) using anti-myc antibodies. Bound proteins were separated by SDS–PAGE and analysed by western blotting using antibodies against the FLAG and myc tags. (c) The same experimental set-up as in (b), except that different FLAG-STIL truncations (depicted in (d)) were co-expressed with myc-βTrCP. (d) Schematic depicting the structures of FLAG-STIL WT and FLAG-STIL truncations used in (c). (e) Same experimental set-up as in (b), except that FLAG-STIL versions carrying mutations in the DSG motif (DAG, AAA) were co-expressed with myc-βTrCP. (f) HEK 293T cells were transfected with either FLAG-STIL WT, FLAG-STIL DAG or FLAG-STIL AAA for 36 h and subsequently treated with either DMSO (control) or MLN4924 for 24 h. Cell lysates were separated by SDS–PAGE and analysed by western blotting using antibodies against the FLAG tag and Aurora A (positive control illustrating effect of MLN4924 treatment). α-tubulin was analysed as loading control. The histogram at the bottom depicts the ratios (MLN4924 treatment relative to DMSO) of STIL WT, STIL DAG and STIL AAA levels, as measured by western blotting in three independent experiments.