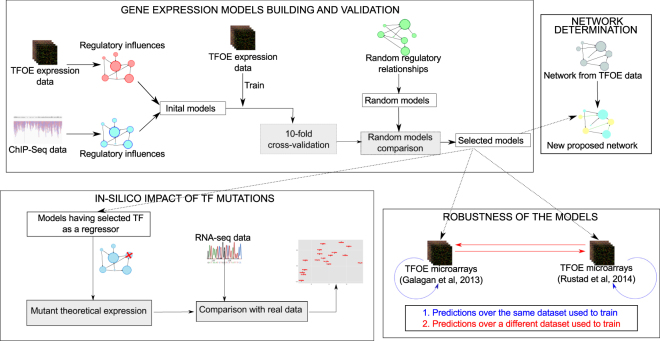
Figure 1.
Workflow to build and validate gene expression computational models. Initial models were derived from the regulatory relationships derived from the TFOE and ChIP-Seq data. They were trained with the data set from Rustad et al.12 (see Materials and Methods for more details about the different data sets used). These initial models went through a 10-fold cross-validation process, to discard low accuracy models. The remaining ones were compared with random models. Those showing better performance than the random models were selected as final models. These models were used to (i) cross-check the robustness of the models, (ii) derive a new regulatory network, and (iii) predict the impact of a TF deletion in silico.