Figure 3.
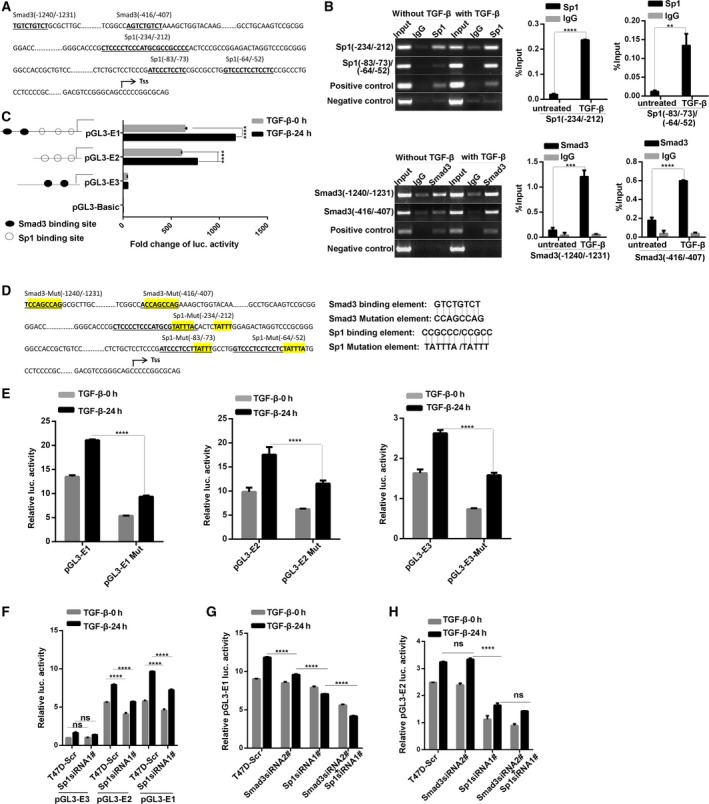
Sp1 and Smad3 bind to the EGFR promoter region and regulate the transcriptional activity of EGFR. (A) Schematic diagram depicting the regulatory sequences of the EGFR promoter region. Three putative binding sites for the transcription factor Sp1 span from −234 to −212, −83 to −73, and −64 to −52 relative to the transcription start site (TSS). The regions between −1240 to −1231 and −416 to −407 were sequences containing putative Smad3‐binding sites. (B) Chromatin immunoprecipitation assay showed that Sp1 and Smad3 specifically immunoprecipitated with EGFR promoter region. T47D cells were pretreated with or without TGF‐β for 6 h, and then, the cell lysates were subjected to chromatin immunoprecipitation using anti‐Sp1 and anti‐Smad3 antibodies. Anti‐RNA polymerase II antibody was used as a positive control. The sequences at −234 to −212, −83 to −73, and −64 to −52 with three Sp1 putative binding sites were specifically immunoprecipitated with anti‐Sp1 antibody. The binding capacity of Sp1 to the EGFR promoter was notably higher in the TGF‐β‐treated group than in the control group. The sequences at −1240 to −1231 and −416 to −407 with two Smad3 putative binding sites were specifically immunoprecipitated with anti‐Smad3 antibody, and TGF‐β treatment significantly enhanced the binding capacity of Smad3 to the EGFR promoter. Right panel, real‐time PCR analysis of the ChIP fragments. Results were analyzed by a percentage of input DNA from TGF‐β‐treated group and the control group. Statistical analysis was carried out by two‐way ANOVA (**P < 0.01, ***P < 0.001). (C) Dual‐luciferase reporter assay showed that the luciferase activity from vectors containing Sp1‐binding sites was stronger than that of other plasmids. TGF‐β treatment can also induce an apparent increase in the luciferase activation from vectors containing Sp1 sequences. Statistical analysis was carried out by two‐way ANOVA (****P < 0.0001). (D) Schematic diagram depicting the regulatory mutation sequences of the EGFR promoter region. The Sp1‐ and Smad3‐binding elements in the pGL3‐E1, E2, and E3 vectors were mutated by PCR‐mediated site‐directed mutagenesis. (E) The luciferase activities of pGL3‐E1/E2/E3 mutants were significantly weaker than those of their corresponding wild‐type plasmids. Statistical analysis was carried out by two‐way ANOVA (****P < 0.0001) (F) Knockdown of Sp1 inhibits the basal and TGF‐β‐induced luciferase activity in pGL3‐E1‐ and pGL3‐E2‐transfected cells, which contain the three putative Sp1‐binding sites. (G,H) Luciferase reporter assay showed that both Sp1 and Smad3 regulate TGF‐β‐induced EGFR transcriptional activation. Downregulation of Smad3 moderately inhibited TGF‐β‐induced luciferase activity in pGL3‐E1‐transfected cells but not in pGL3‐E2‐transfected cells, whereas double knockdown of Sp1 and Smad3 showed the strongest inhibition of luciferase activity among the other cases. All the data were presented as mean ± SD, and the experiments were repeated three times. Statistical analysis was carried out by two‐way ANOVA (****P < 0.0001).
