Figure 1.
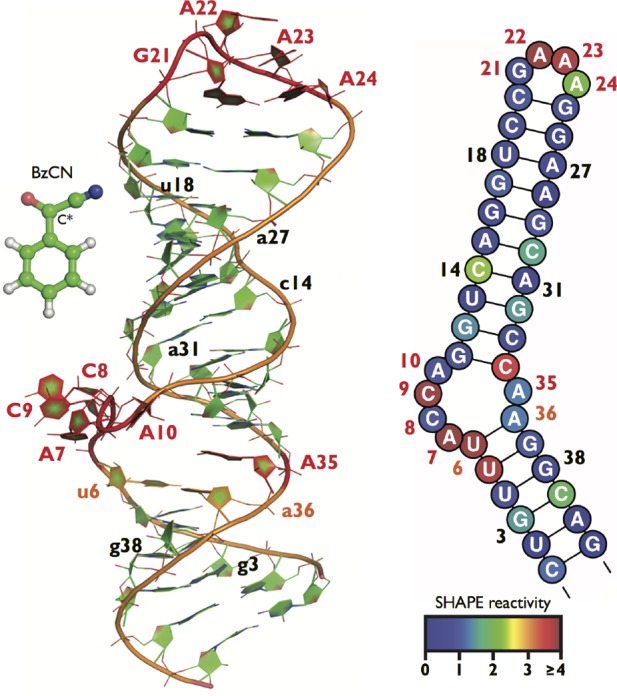
Tertiary and secondary structures of SRP–RNA motif. The left panel shows the tertiary structure used as starting point for MD simulations (PDB ID 1DUL).39 Nucleotides from GNRA tetraloop, first and last noncanonical base pairs from the symmetric loop, all nucleotides from the asymmetric internal loop, and U6-A36 closing base pair from the asymmetric loop are labeled (unpaired nucleotides in capital letters). Structural model of small BzCN reagent is displayed (not to scale) on the left side with the reactive carbon (C*) highlighted. The right panel shows the corresponding secondary structure model, where the coloring scheme for each nucleotide is based on experimentally measured SHAPE reactivity.43
