Figure 1.
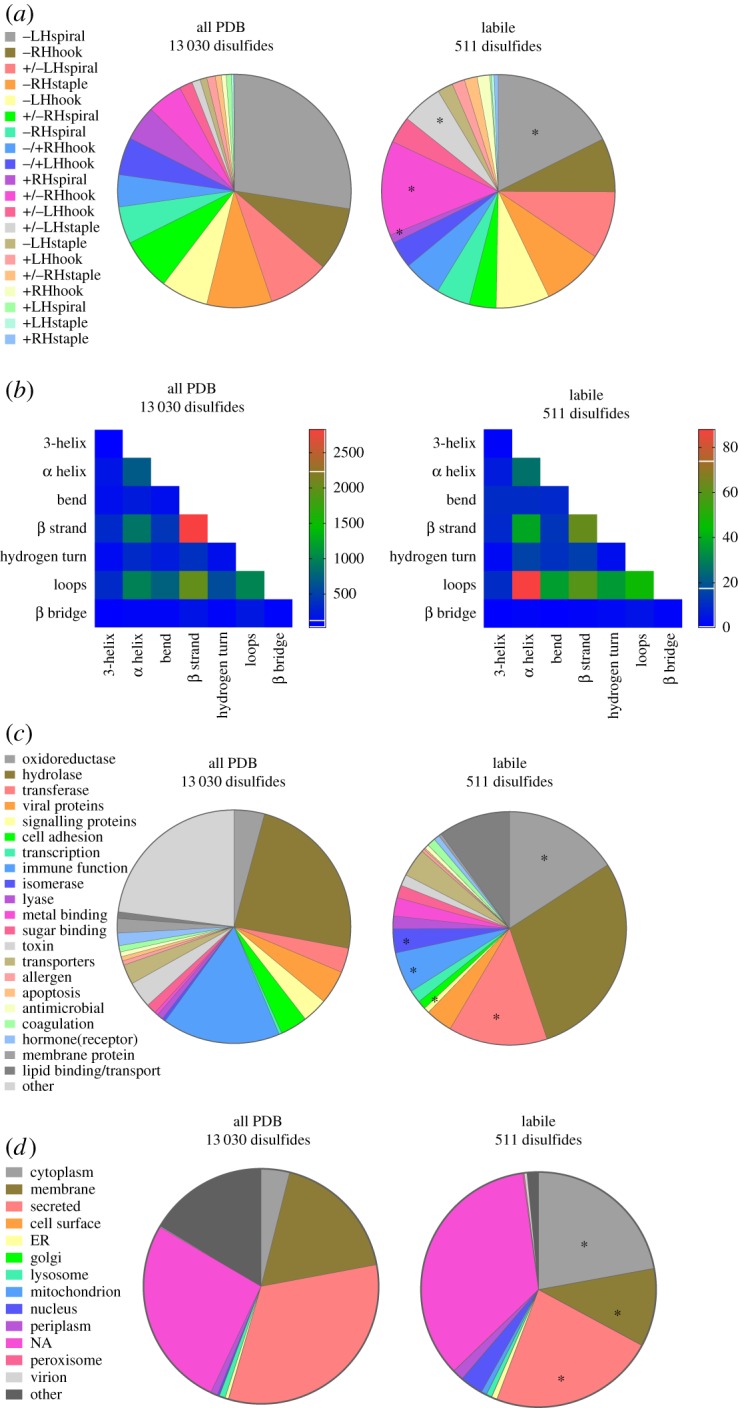
Structural and functional features of the labile disulfide bonds. (a) Distribution of the 20 disulfide bond configurations in unique disulfide bonds in PDB protein X-ray structures (13 030 disulfides, electronic supplementary material, table S1) and in labile bonds (511 disulfides, electronic supplementary material, table S2). Compared with the total PDB, the labile bonds are enriched in +/–RHhook and +/–LHstaple bonds (χ2 test, p < 0.0001) and have relatively fewer –LHspiral and +RHspiral bonds (χ2 test, p < 0.0001) (indicated by *). (b) Heatmap displaying the frequency of the secondary structures linked by disulfide bonds in all PDB protein structures and by labile disulfide bonds. There is enrichment of disulfides linking α-helices and loops in labile disulfide bonds (χ2 test, p < 0.0001). (c) Distribution of the functional classification of all proteins containing disulfide bonds and proteins containing labile disulfide bonds. Compared to the total PDB, there was a significant increase in oxidoreductases, transferases and isomerases (indicated by *). A significant decrease in disulfide bonds in proteins involved in signalling and immune function was observed (χ2 test, p < 0.0001). (d) Subcellular localization of all proteins containing disulfide bonds and proteins containing labile disulfide bonds. Compared to the total PDB, a significant increase in cytoplasmic proteins, as well as a decrease in membrane associated and secreted proteins was observed (χ2 test, p < 0.0001) (indicated by *).
