Figure 6.
Impaired Pluripotency and Differentiation in Murine Embryonic Stem Cells with Low GSH Levels
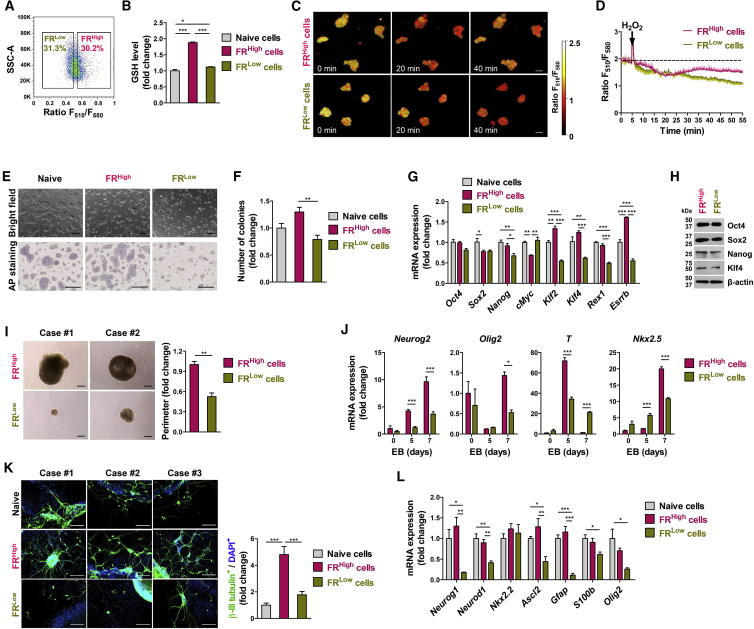
(A) Sorting of murine ESCs (mESCs) according to the F510/F580 ratio (FR) into FRHigh and FRLow populations.
(B) Luminescence-based quantification of GSH in cell lysates (n = 4 independent biological replicates).
(C and D) Representative pseudo-colored confocal images of the FR (C) and the comparison of their FR changes (n = 4 cells; D) in FRHigh and FRLow mESCs, following treatment with 100 μM H2O2. Scale bars, 10 μm.
(E) Representative pictures for bright-field (upper panel) and alkaline phosphatase (AP) staining (lower panel). Scale bars, 200 μm.
(F–H) Clonogenic capacity in limiting dilution (n = 5 independent biological replicates; F), qPCR (n = 4 independent biological replicates; G), and western blot (H) analyses in FRHigh and FRLow mESCs and in unsorted control (naive) cells.
(I) Representative pictures (left) and quantitation (n = 6 independent biological replicates; right) for size of 5-day embryoid body (EB) formed from FRHigh and FRLow mESCs. Scale bars, 200 μm.
(J) qPCR analyses for lineage-specific genes (Neurog2, Olig2, T, and Nkx2.5) in EBs at the indicated day (n = 4 independent biological replicates).
(K and L) Immunostaining of βIII-tubulin+ neuron cells (green; K) qPCR analyses of neural lineage markers (Neurog1, Neurod1, Nkx2.2, Ascl2, Gfap, S100b, and Olig2; n = 4 independent biological replicates; L) during ESC differentiation. Nuclei were counterstained with DAPI (blue; K). The βIII-tubulin+ neuron cells were quantitated by their frequency in total DAPI+ cells (n = 5 independent biological replicates; K, right panel). Scale bars, 100 μm.
Values represent mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S6.