Figure 5. NBR1 cooperates with p62.
-
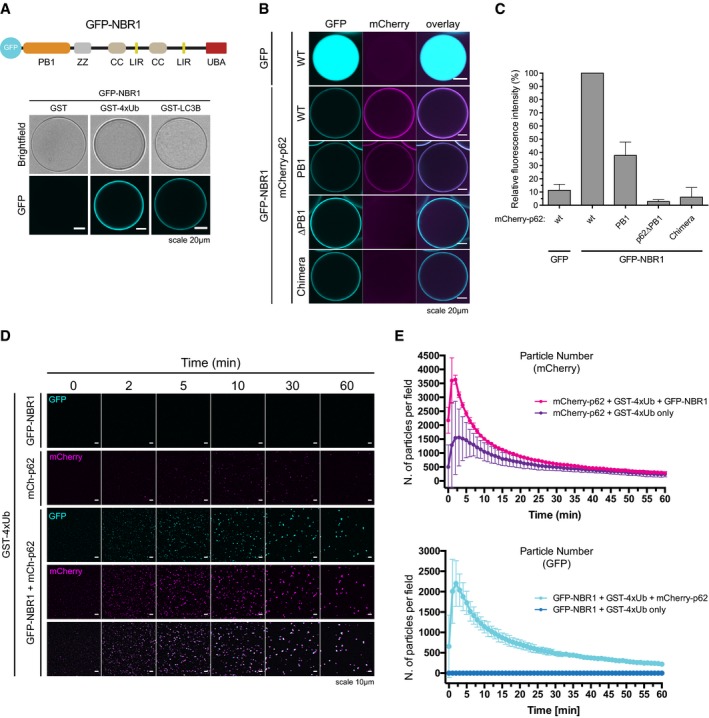
ATop: schematic representation of the domain architecture of the GFP‐NBR1 construct. PB1: Phox and Bem1p domain, ZZ: Zinc finger, CC: coiled coil, LIR: LC3‐interacting region, UBA: ubiquitin‐associated domain. Bottom: GFP‐NBR1 was recruited to glutathione beads coated with the indicated GST fusion proteins and imaged by spinning disk microscopy. Representative micrographs of at least 70 beads imaged per sample from two independent experiments.
-
B, CmCherry‐p62 variants were recruited to GFP‐ or GFP‐NBR1‐coated beads and imaged as in (A). (B) Representative micrographs of at least 70 beads per sample from three independent replicates. (C) Quantification of the data shown in (B). Data were normalized to p62 wild‐type binding to GFP‐NBR1 within each replicate. Averages and SDs of three independent experiments are shown.
-
D, E2 μM mCherry‐p62 and/or GFP‐NBR1 were incubated in the presence of 5 μM GST‐4xUb. Representative micrographs (D) and quantification of particle number for each channel (E) are shown. Averages and SDs from at least three independent experiments are shown.
