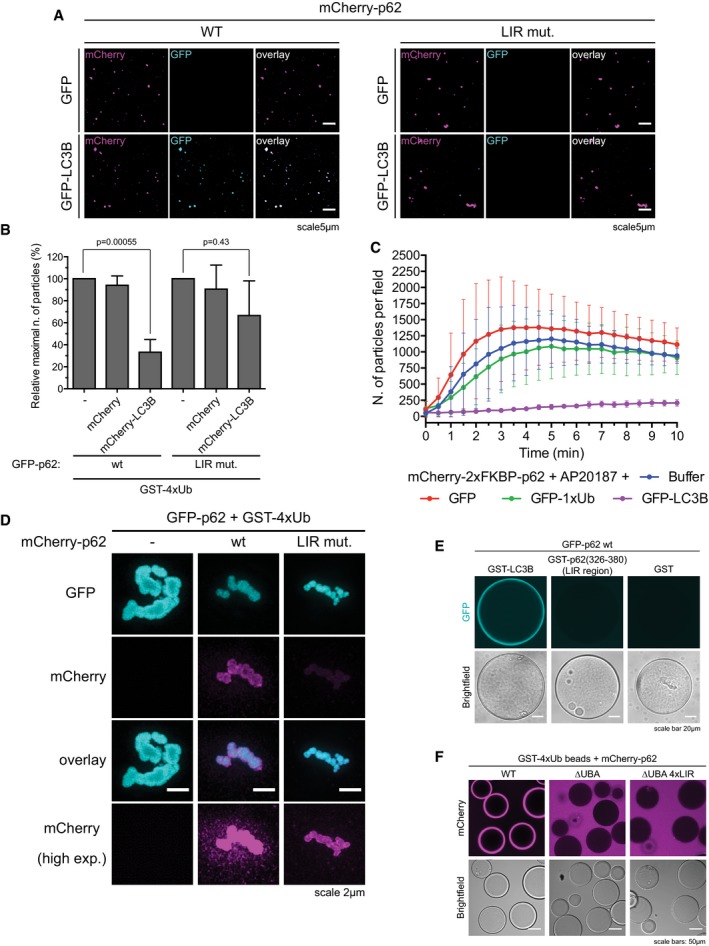
Figure EV6. LIR‐mediated cross talk between cluster formation and the autophagy machinery.
-
ARepresentative micrographs of clusters formed with 1 μM mCherry‐p62 WT or LIR mutant and 20 μM GST‐4xUb for 10 min, followed by the addition of 1 μM GFP or GFP‐LC3B immediately prior to imaging. Clusters deposited on the bottom of the plate were imaged by spinning disk microscopy at 63× magnification.
-
BGFP‐p62‐ and GST‐4xUb‐containing clusters were formed in the presence of pre‐mixed 20 μM mCherry or mCherry‐LC3B employing p62 wild type or the LIR mutant. The maximal number of particles per field formed per sample was taken as readout. Data were normalized to the samples containing p62 and GST‐4xUb only for both p62 wild type and the LIR mutant. Averages and SDs from three independent experiments are shown. P‐values were calculated with a two‐tailed unpaired Student's t‐test.
-
CQuantification of cluster formation reactions conducted with 2 μM mCherry‐2xFKBP‐p62 in the presence of 5 μM GST‐4xUb and 20 μM of the indicated proteins. Averages and SDs from three independent replicates are shown.
-
DRepresentative SIM micrographs of p62 clusters formed with the indicated proteins. GFP‐p62‐containing clusters were formed for 30 min, and then, the respective mCherry‐p62 mutants were added and incubated for another 30 min. Representative images of at least 10 clusters per sample are shown.
-
E, FThe indicated p62 variants were recruited to glutathione beads coated with the indicated GST‐tagged proteins and imaged under a spinning disk microscope (E) or a conventional confocal microscope (F). Representative pictures of two independent experiments per condition are shown.
