Summary
The heterochromatin protein 1 (HP1) family is involved in various functions with maintenance of chromatin structure. During murine somatic cell reprogramming, we find that early depletion of HP1γ reduces the generation of induced pluripotent stem cells, while late depletion enhances the process, with a concomitant change from a centromeric to nucleoplasmic localization and elongation-associated histone H3.3 enrichment. Depletion of heterochromatin anchoring protein SENP7 increased reprogramming efficiency to a similar extent as HP1γ, indicating the importance of HP1γ release from chromatin for pluripotency acquisition. HP1γ interacted with OCT4 and DPPA4 in HP1α and HP1β knockouts and in H3K9 methylation depleted H3K9M embryonic stem cell (ESC) lines. HP1α and HP1γ complexes in ESCs differed in association with histones, the histone chaperone CAF1 complex, and specific components of chromatin-modifying complexes such as DPY30, implying distinct functional contributions. Taken together, our results reveal the complex contribution of the HP1 proteins to pluripotency.
Keywords: HP1α knockout, HP1β knockout, HP1γ knockout, pluripotency, iPSC, reprogramming, Dppa4, Senp7, H3.3, H3K9M
Highlights
-
•
Release of HP1γ from anchoring by Senp7 increases reprogramming efficiency
-
•
HP1γ switches enrichment from histone H1 to histone H3.3 in pluripotent cells
-
•
HP1γ interacts with OCT4 and DPPA4 independent of HP1α, HP1β, and H3K9 methylation
-
•
Proteomic characterization of HP1 protein family in pluripotent cells
In this article, Sridharan and colleagues perform proteomic characterization of the heterochromatin protein 1 (HP1) family in reprogramming and pluripotency. Depletion of HP1γ or its interacting partner SENP7, which anchors it to heterochromatin, increased iPSC generation. In pluripotent cells, HP1γ is highly nucleoplasmic and enriched with histone H3.3. HP1γ interacts with OCT4 and DPPA4 independent of HP1α, HP1β, and H3K9 methylation. HP1α and HP1γ differ in association with specific components of chromatin-modifying complexes such as DPY30.
Introduction
Embryonic stem cells (ESCs) have the unique ability to self-renew indefinitely and differentiate into several cell types in response to the appropriate stimuli. This remarkable plasticity is correlated with a chromatin structure that is less enriched for compacted heterochromatic DNA and has a higher mobility of chromatin-associated proteins such as heterochromatin protein 1 (HP1) alpha than somatic cells (Fussner et al., 2011, Meshorer and Misteli, 2006, Meshorer et al., 2006, Shchuka et al., 2015).
In mammals, the HP1 family consists of three proteins: HP1α, HP1β, and HP1γ, which have a highly conserved chromodomain that binds to histone H3 lysine 9 methylation (H3K9me), a transcriptionally repressive chromatin modification, and a chromoshadow domain that is involved in protein-protein interactions (Bannister et al., 2001, Smothers and Henikoff, 2001). While the HP1 proteins participate in diverse cellular processes in somatic cells, such as nucleating regions of repression (Nielsen et al., 1999), their role in pluripotency is poorly understood. The depletion of the HP1 proteins in mouse ESCs does not lead to an extensive change in mRNA or repetitive element expression (Bulut-Karslioglu et al., 2014, Maksakova et al., 2013, Mattout et al., 2015, Sridharan et al., 2013), but may affect splicing in conjunction with DNA methylation (Smallwood et al., 2012, Yearim et al., 2015). In mouse ESCs, knockout of HP1β impaired pluripotency and mesodermal differentiation (Mattout et al., 2015), while HP1γ depletion leads to endodermal and neural differentiation defects (Caillier et al., 2010, Huang et al., 2017). The HP1β knockout mouse is perinatally lethal due to neural defects, whereas the HP1γ knockouts, although viable, exhibit severe infertility (Aucott et al., 2008, Brown et al., 2010, Takada et al., 2011).
Reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) that resemble ESCs (Takahashi and Yamanaka, 2006) provides an opportunity to examine how each of these proteins can influence the acquisition of pluripotency. We have previously found that depleting HP1γ caused a greater increase in murine reprogramming efficiency than depleting HP1α or HP1β (Sridharan et al., 2013). To investigate differing functional roles for these proteins, we performed immunofluorescence assays and found that, unlike HP1α, the majority of HP1γ was nucleoplasmic upon reaching the iPSC state. Concomitant with this change in localization, there was an increase in iPSCs obtained with a knockdown of HP1γ. Proteomics characterization of HP1γ complexes from mouse ESCs and reprogramming intermediates called partially reprogrammed cells (pre-iPSCs) revealed enrichment with elongation-associated histone H3.3 and linker histone H1 respectively. The depletion of SUMO protease Senp7, which anchors HP1 to heterochromatin (Maison et al., 2011, Maison et al., 2012, Romeo et al., 2015) and is more enriched with HP1γ in pre-iPSCs, increases reprogramming efficiency. In ESCs, we identified interactions of HP1γ with pluripotency-related proteins DPPA4 and OCT4. Although all three HP1 proteins interacted with chromatin-associated complexes, HP1γ was more enriched for certain components, such as DPY30 of the MLL complex, suggesting that subunit composition may aid in targeting different HP1 proteins. We found several post-translational modifications of the HP1 proteins in ESCs. These results provide insights into how the interactions of these important heterochromatin proteins influence their role in pluripotency.
Results
HP1γ Localization Changes with Temporal Requirement in Reprogramming
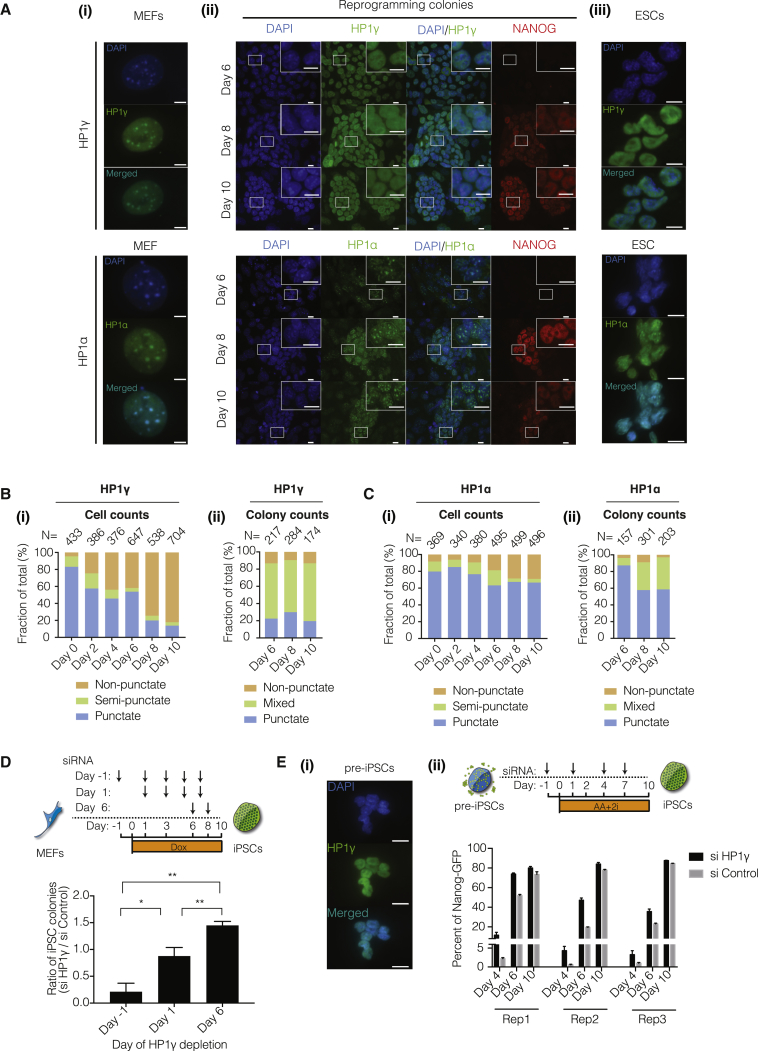
In order to determine how the localization of HP1γ changed during the generation of iPSCs, we initiated reprogramming in mouse embryonic fibroblasts (MEFs). Similar to HP1α, HP1γ colocalized with punctate DAPI-intense heterochromatin in greater than 80% of MEFs (Figures 1A, 1B, 1C, and S1A). In ESCs, HP1γ was almost completely nucleoplasmic (91.9%), in contrast with colocalization of HP1α with the DAPI foci (70.1%) (Figures 1A and S1B). The transcript levels of HP1γ plateaued at day 6 of reprogramming, similar to those of HP1α and HP1β (Figure S1B). By day 6, the reprogramming cells had clustered to form colonies that did not express pluripotency factor Nanog, but HP1γ was already in a more nucleoplasmic configuration. This suggests that HP1γ relocalization precedes iPSC formation. By day 8 of reprogramming, ∼70% of the Nanog-positive iPSC colonies had a mostly nucleoplasmic distribution of HP1γ (Figures 1A, 1B, and S1C). In contrast, ∼60% of iPSC colonies had punctate localization for HP1α (Figures 1A, 1C, and S1C). To observe whether HP1γ that was nucleoplasmic returned to a punctate localization, we performed time-lapse live-cell imaging. Exogenously expressed GFP-tagged HP1γ in MEFs showed a punctate localization, similar to endogenous HP1γ (Figure S1D). Upon reprogramming, the GFP-tagged HP1γ changed in localization from a punctate to nucleoplasmic location. Importantly, nucleoplasmic HP1γ retained such localization during reprogramming (Figure S1D and Movie S1).
Figure 1.
Differential Localization and Function of HP1γ during Reprogramming
(A) Immunofluorescence for HP1γ (top panel) and HP1α (bottom panel) in (i) mouse embryonic fibroblasts (MEFs), (ii) at different time points in reprogramming populations, and (iii) embryonic stem cells (ESCs). Scale bar, 10 μm. Inset: zoomed image of a region (white box). Inset scale bar, 10 μm.
(B) (i) Cell counts of punctate, non-punctate, and semi-punctate HP1γ localization in reprogramming MEFs, plotted as a fraction of the total cells counted, N. Semi-punctate refers to a generally more nucleoplasmic localization, but still exhibiting <3 punctate spots. (ii) Colony counts of punctate, non-punctate, and mixed HP1γ localization in reprogramming colonies, plotted as a fraction of the total colonies counted, N.
(C) (i) Cell counts of punctate, non-punctate, and semi-punctate HP1α localization in reprogramming MEFs plotted as a fraction of the total cells counted, N. Semi-punctate refers to a generally more nucleoplasmic localization, but still exhibiting <3 punctate spots. (ii) Colony counts of punctate, non-punctate, and mixed HP1α localization in reprogramming colonies, plotted as a fraction of the total colonies counted, N.
(D) Top: scheme of experiment. Doxycycline (Dox) induction of reprogramming factors on day 0, small interfering RNA (siRNA) transfection on days indicated by arrows. Nanog-positive iPSCs colonies counted on day 10. Bottom: ratio of iPSC colonies formed upon depletion of HP1γ over non-targeting control show differences between each depletion starting point. Error bars represent SDs of three independent reprogramming experiments. ∗p < 0.05, ∗∗p < 0.01 assessed by Student's t test.
(E) (i) Immunofluorescence of HP1γ in partially reprogrammed iPSCs (pre-iPSCs). Scale bar, 10 μm. (ii) Top: scheme of experiment. Pre-iPSCs treated with AA + 2i: AA, ascorbic acid; 2i, MAPK inhibitor (PD-0325901) + GSK inhibitor (CHIR-99021). Cells were transfected with siRNA on indicated days. Bottom: percentage of Nanog-GFP cells upon depletion of HP1γ or non-targeting control determined by flow cytometry. Error bars represent SDs of two independent wells from the same experiment. Data from three independent conversion experiments are presented.
See also Figures S1 and S2, and Movie S1.
To determine whether this temporal change in localization had any functional effects, we performed a time course of HP1γ depletion by using small interfering RNA (siRNA)-mediated knockdown (Figure S2A) during reprogramming (Figures 1D and S1C). Remarkably, when HP1γ was depleted early in reprogramming (day −1), efficiency was reduced compared with depletion later (day 6) when efficiency was increased (Figure 1D). The early reduction in colony number was not due to reduced cell numbers during reprogramming of HP1γ-depleted MEFs (Figure S2B). Since the reprogramming population represents a heterogeneous mixture of cells that could respond to HP1γ depletion differently, we used a pre-iPSCs system. Pre-iPSCs are clonal transcriptional intermediates of the reprogramming process that have not activated the pluripotency network and can convert to iPSCs at low efficiency with HP1γ depletion (Sridharan et al., 2013, Sridharan et al., 2009). HP1γ is largely nucleoplasmic in pre-iPSC (Figure 1E). We have recently developed a method where the combination of ascorbic acid and 2i (mitogen-activated protein kinase [MAPK] and glycogen synthase kinase [GSK] inhibition) converts pre-iPSCs to iPSCs at 80% efficiency (Tran et al., 2015). Given the robustness of this conversion, very few modulators are expected to have a further effect. We depleted HP1γ in this high-efficiency system and found that reprogramming kinetics increased, with a significant number of Nanog-GFP reporter-positive cells appearing by day 4 (Figures 1E and S2A). Thus, after the change in localization of HP1γ from DAPI associated to more nucleoplasmic, depleting the levels of HP1γ increases reprogramming efficiency.
High Salt and Nuclease Digestion Release Different Kinds of HP1γ Complexes
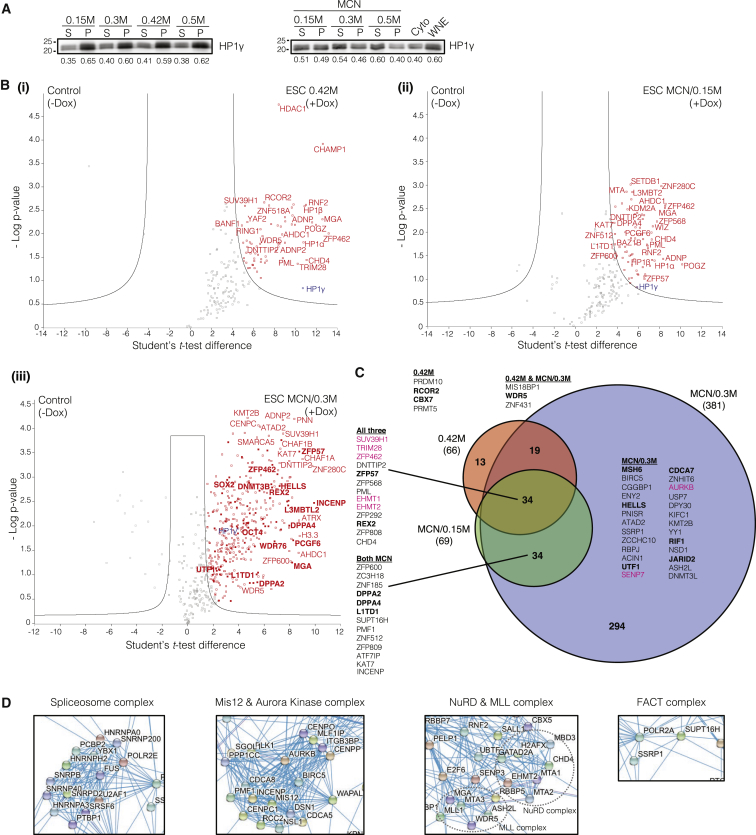
To investigate these changing requirements for HP1γ, we decided to identify interactors of HP1γ that correlate with the cell fate change to a pluripotent state. In general, transcription factors are soluble by extraction with 0.42 M salt (Dignam et al., 1983). We extracted nuclear proteins from ESCs at increasing salt concentrations from 0.15 M to 0.42 M and with micrococcal nuclease (MCN) digestion to release nucleoplasmic or chromatin-bound proteins respectively. In parallel with the increasing amount of soluble proteins proportional to the salt concentration in isolated nuclei (Figure S2C), immunoblotting for HP1γ revealed that 59% remained in the insoluble pellet fraction under 0.42 M salt conditions, while 46% of HP1γ remained in the pellet at 0.3 M salt in MCN conditions (Figure 2A). Thus, the more nucleoplasmic localization of HP1γ in pluripotent cells does not translate to complete extraction even at 0.5 M salt.
Figure 2.
Compartmentalization of HP1γ Complexes in ESCs
(A) Immunoblot of HP1γ extracted from ESCs with increasing salt concentrations (left), or micrococcal nuclease (MCN)-mediated extraction with increasing salt concentrations (right). S, supernatant; P, pellet; WNE, whole nuclear extract; Cyto, cytoplasmic. Bottom: numbers indicate quantified fraction of HP1γ from S and P with total S + P = 1.
(B) HP1γ-interacting proteins enriched in different extraction methods. Volcano plots show proteins enriched in FLAG-HP1γ of ESC (+Dox) extracted with (i) 0.42 M KCl, (ii) MCN with 0.15 M NaCl, or (iii) MCN with 0.3 M NaCl against uninduced samples (-Dox) as control. Significantly enriched proteins, red; bait, blue; significantly enriched proteins that are also expressed >10-fold higher in ESC compared with MEF, bold red.
(C) Venn diagram showing overlaps of enriched proteins between the three extraction conditions as in (B). Known interactors of HP1γ, magenta; previously unidentified interactors of HP1γ, black; previously unidentified interactors of HP1γ that are also expressed 10-fold higher in ESC, bold black.
(D) Complexes enriched in FLAG-HP1γ IP-MS of MCN with 0.3 M NaCl overlaid on the STRING Network database.
ESC lines with N-terminal FLAG-epitope-tagged HP1γ at a single genomic location were generated and induced to mimic endogenous levels (Figure S2D). From these ESC lines, anti-FLAG immunoprecipitation followed by mass spectrometry (IP-MS) was performed in nuclear extracts prepared with different salt and enzymatic conditions (Experimental Procedures, Figures 2B and 2C, Table S1). The 0.42 M salt and MCN + 0.15 M salt yielded ∼66 interacting proteins compared with 381 interactors in the MCN + 0.3 M salt condition (Figure 2C). Components of the centromere proximal complex (e.g., INCENP), histone-modifying enzymes that regulate centromere assembly (e.g., KAT7) (Ohzeki et al., 2016), and several zinc finger proteins (e.g., ZFP600 and ZNF512) were found in both MCN + salt conditions. HP1γ interacted with PRMT5, an arginine methyltransferase, and CBX7, a polycomb-related protein, exclusively in 0.42 M salt extraction. In both the high-salt conditions, WDR5, a protein in the MLL complex, was enriched (Figure 2C). Thus HP1γ-associated protein complexes can be partitioned in biochemically distinct environments.
To determine the pluripotent-specific interactions of HP1γ in the MCN + 0.3 M salt condition, we identified proteins whose gene expression level was 10-fold greater in ESCs compared with MEFs (Sridharan et al., 2009). We found the ESC-specific protein REX2, an uncharacterized zinc-finger-domain-containing protein, two poorly studied ESC-specific proteins, DPPA2 and DPPA4, and L1TD1, a protein involved in RNA post-transcriptional processes as HP1γ interactors (Figures 2B and 2C and Table S1). ESC-enriched transcription factors including UTF1, OCT4, and ESRRB, but not NANOG, were also detected in this pull-down (Figure 2B). We also found interactions with the MIS12-containing DNA replication complex, the chromatin remodeling NuRD complex, and FACT complexes (Figure 2D). We next applied these extraction conditions to pre-iPSCs for comparative profiling.
Differential Association of HP1γ Complexes in Pre-iPSCs
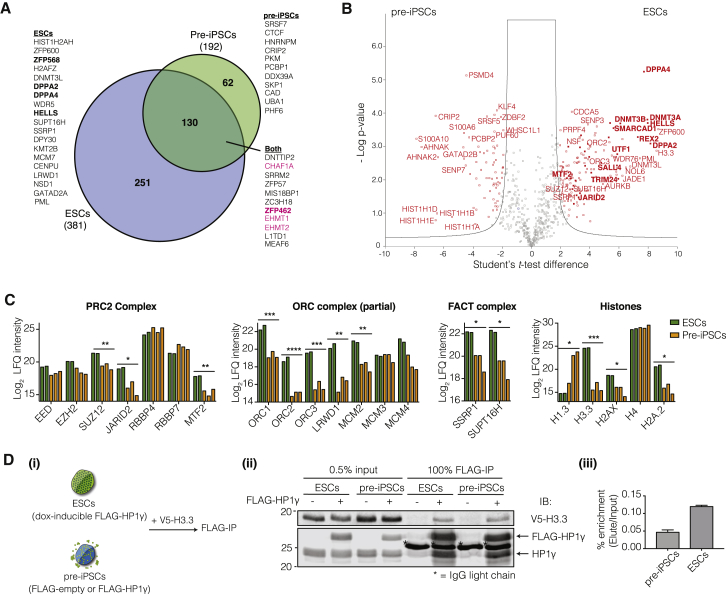
We generated FLAG-tagged HP1γ pre-iPSC lines with expression equivalent to endogenous HP1γ levels (Figure S3A) and performed IP-MS in MCN + 0.3 M salt conditions. Similar to ESCs, interactions of HP1γ with the histone chaperone CAF1 complex and H3K9 methyltransferases were detected in pre-iPSCs (Figures 3A and S3B and Table S1). Certain pluripotency-specific interactions of HP1γ, such as with L1TD1 and ZFP57, were already detected at the pre-iPSCs stage, likely because their expression had already reached ESCs levels (Figures 3A, 3B, and S3B).
Figure 3.
Differential Enrichment of HP1γ-Interacting Proteins in Pre-iPSCs Compared with ESCs
(A) Venn diagram showing overlaps of enriched HP1γ interactors from individual analyses in ESCs (as in Figure 2B) and pre-iPSC (as in Figure S3B). Known interactors of HP1, magenta; previously unidentified interactors of HP1, black; interactors of HP1 that are also expressed >5-fold higher in ESC compared to pre-iPSC, bold black.
(B) Volcano plot showing quantitative comparison of FLAG-HP1γ IP-MS in ESC versus pre-iPSC. Significantly enriched proteins, red; significantly enriched proteins that are also expressed >5-fold higher in ESC compared to pre-iPSC, bold red.
(C) Bar graphs show differential enrichment of HP1γ-interacting proteins as part of complexes between ESCs and pre-iPSCs. Data from two or three independent immunoprecipitation experiments for ESC and pre-iPSC, respectively, are presented. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001 assessed by Student's t test.
(D) (i) Scheme of experiment: ESCs with dox-inducible FLAG-HP1γ or pre-iPSCs with either FLAG-empty or FLAG-HP1γ were transduced with V5-tagged H3.3 lentivirus followed by immunoprecipitation with FLAG antibody (FLAG-IP). (ii) Immunoprecipitation (IP) with FLAG antibody with immunoblot for V5 and HP1γ. Asterisk (∗) marks immunoglobulin G (IgG) light chain (25 kDa). (iii) Quantitation of percentage enrichment (elute/input) for V5-H3.3 in pre-iPSC and ESC. Error bars represent SDs of two independent immunoprecipitation experiments.
We exploited the sensitivity of the mass spectrometry approach to observe subtle changes in composition of chromatin-related complexes that are unlikely to be detected by western blots. Among the PRC2 members, JARID2 and MTF2 are less enriched in pre-iPSCs and the ORC complex is less abundant in its interaction with HP1γ (Figures 3B and 3C). The GATAD2B (P66β) component of the NuRD complex is more enriched in pre-iPSCs compared with ESCs (Figures 3B and 3C). By gene expression levels all these components are similarly expressed in pre-iPSC and ESCs (Sridharan et al., 2009). The FACT complex, which disrupts the H2A-H2B dimer to allow RNA polymerase to read through and promotes transcription elongation, is less enriched in pre-iPSCs than in ESCs (Figure 3C).
A striking difference between the two cell types was in the association of HP1γ with specific histones. In pre-iPSCs, there was a significant enrichment of HP1γ with the histone H1 proteins that play a role in chromatin compaction (Figures 3B and 3C). There was also a lower enrichment of histone H3.3, which is associated with transcription elongation and the DNA-repair-associated H2AX in pre-iPSCs (Figures 3B and 3C) (Ahmad and Henikoff, 2002, McKittrick et al., 2004). To confirm this differential enrichment, we generated V5-tagged H3.3 ESC and pre-iPSC cell lines (Figure 3D). By performing immunoprecipitation for FLAG-HP1γ we were able to quantify the levels of H3.3 that was specifically bound in each cell type. There was a 2.5-fold increase in the amount of H3.3 association in ESCs compared with pre-iPSCs, verifying the results from mass spectrometry (Figure 3D). Taken together, the increased HP1γ interaction with H3.3 and the FACT complex is likely to promote transcriptional elongation in ESCs.
SENP7 May Aid in Release of HP1γ in Pre-iPSCs
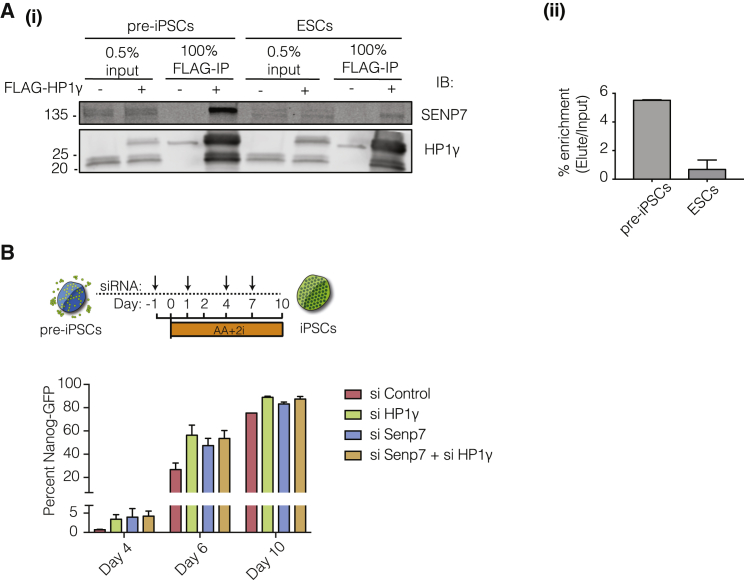
HP1α undergoes a SUMOylation-deSUMOylation cycle for recruitment to centromeric regions (Maison et al., 2012, Maison et al., 2011, Romeo et al., 2015). Initially SUMO-1 adds SUMO-groups to HP1α, leading to centromeric recruitment followed by deSUMOylation by SENP7, leading to centromeric anchoring. There was a greater association of SENP7 with HP1γ in pre-iPSCs compared with ESCs (Figure 3B). Quantitative immunoprecipitation revealed a 5-fold greater interaction of SENP7 with HP1γ in pre-iPSCs compared with ESCs (Figure 4A).
Figure 4.
Differential Enrichment of SENP7 in Pre-iPSCs Compared with ESCs
(A) (i) IP with FLAG antibody in pre-iPSCs and ESCs with immunoblot for SENP7 and HP1γ. (ii) Quantitation of percentage enrichment (elute/input) for SENP7 in pre-iPSCs and ESCs. Error bars represent SDs of two independent immunoprecipitation experiments.
(B) Percentage of Nanog-GFP cells in a pre-iPSC conversion experiment upon depletion of Senp7, HP1γ, both Senp7 and HP1γ, or non-targeting control. Error bars represent SDs of two independent conversion experiments.
See also Figure S4.
To determine whether the differential association of SENP7 with HP1γ had a functional consequence for pluripotency, we depleted Senp7 in pre-iPSCs during reprogramming (Figure S4A). Using the same high-efficiency system for conversion of pre-iPSCs as in Figure 1, we found that the decrease in Senp7 caused an increase in reprogramming efficiency and kinetics, similar to what is observed with depletion of HP1γ (Figure 4B). Interestingly the combined depletion of both Senp7 and HP1γ did not enhance the conversion efficiency to the iPSC state (Figure 4B). This result suggests that both SENP7 and HP1γ act in the same pathway and that reducing the anchoring of HP1γ is important for reaching the pluripotent state.
Pluripotency Factor Association of HP1γ
We chose to interrogate its interaction with pluripotency factors OCT4 and DPPA4 in further detail. DPPA4 is a SAP-domain-containing protein (Maldonado-Saldivia et al., 2006). Both HP1γ and Dppa4 knockout mice have defects in spermatogenesis (Brown et al., 2010, Madan et al., 2009, Nakamura et al., 2011, Takada et al., 2011).
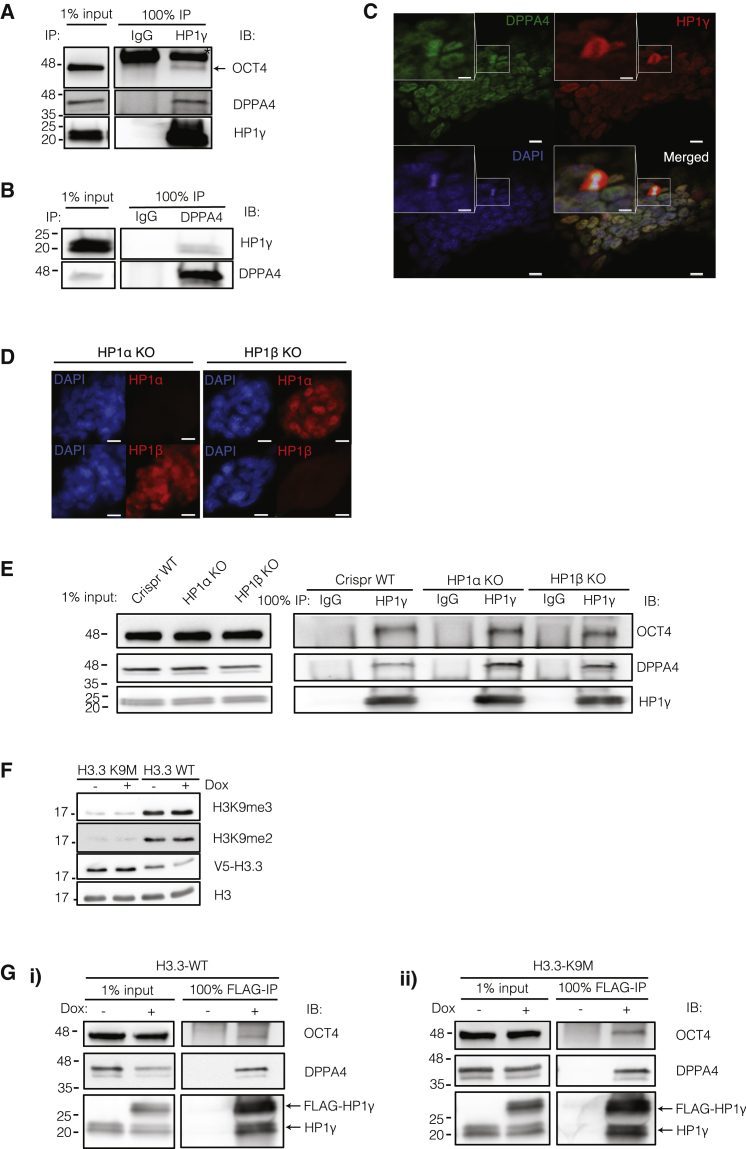
We confirmed the interaction of both endogenous and FLAG-tagged HP1γ with OCT4 and DPPA4 (Figures 5A and S5A). The reciprocal IP using DPPA4 antibody also confirmed HP1γ as a bona fide interactor (Figure 5B). These interactions were retained after immune complexes were washed at 0.5 M salt (Figure S5B) and were specific because other abundant proteins in ESCs, such as DPPA3, were not immunoprecipitated (Figure S5A). These interactions were also retained after Benzonase digestion (Figure S5C), indicating that the interaction was not due to spurious DNA bridging. Interestingly, DPPA4 and HP1γ colocalize in the nucleoplasm of interphase nuclei but not in mitotically dividing cells, as observed by confocal microscopy (Figure 5C). DPPA4 remains tightly localized to DAPI-stained DNA, which may suggest a function in mitotic bookmarking that is independent of its association with HP1γ or at least the mitotically associated Ser-83 phosphorylated form of HP1γ (Leonard et al., 2015).
Figure 5.
Interaction of HP1γ with OCT4 and DPPA4 Is Independent of the Presence of HP1α or HP1β and H3K9 Methylation
(A) Immunoprecipitation (IP) of endogenous HP1γ with immunoblot for OCT4 and DPPA4. Asterisk (∗) marks IgG heavy chain (∼55 kDa).
(B) IP of endogenous DPPA4 with immunoblot for HP1γ.
(C) Confocal immunofluorescence image show regions of colocalization of DPPA4 with HP1γ except in mitotically dividing cells. Scale bar, 10 μm; inset scale bar, 5 μm.
(D) Immunofluorescence of HP1α and HP1β ESC knockout cell lines. Scale bar, 10 μm.
(E) IP of endogenous HP1γ in Crispr wild-type (WT), HP1α KO, and HP1β KO ESC with immunoblot for OCT4 and DPPA4.
(F) Western blot of H3K9me2 and H3K9me3 in FLAG-HP1γ ESC line with V5-tagged H3.3 K9M compared with control V5-tagged H3.3 WT. Cells were treated with doxycycline (Dox) to induce expression of FLAG-HP1γ.
(G) IP of FLAG-HP1γ in (i) H3.3 WT ESC and (ii) H3.3 K9M ESC, with immunoblot for OCT4 and DPPA4.
See also Figure S5.
Since HP1γ forms heterodimers with HP1α and HP1β, we examined whether OCT4 and DPPA4 require heterodimerization to interact with HP1γ. Using CRISPR-based technology we generated knockout ESC lines of HP1α or HP1β individually (Figures 5D and S5D). The levels of HP1γ were not affected in these cell lines and both the knockout cell lines could be propagated efficiently (data not shown). In the context of the HP1α or HP1β knockout (KO), HP1γ interaction with both OCT4 and DPPA4 was retained (Figure 5E).
DPPA4 contains a PxVxL motif that is commonly found in several HP1-interacting proteins (Thiru et al., 2004) and could represent the interaction surface. However, mutation of this motif did not affect the binding of DPPA4 to HP1γ (Figure S5E). Moreover, the interaction between DPPA4 and HP1γ was only observed under the MCN extraction conditions, which release chromatin-bound proteins (Figure 2C). Therefore, we wondered whether the interaction of DPPA4 with HP1γ was due to the embedding of DPPA4 in H3K9 methylation-enriched heterochromatin.
To deplete H3K9 methylation, we generated ESC lines with a stable integration of a histone H3.3 lysine 9 to methionine mutation (H3K9M) (Lewis et al., 2013). This mutant form of H3.3 has been shown to trap the SET-domain-containing histone methyltransferases (Lewis et al., 2013). ESC lines with greater than 90% reduction in H3K9me2 and H3K9me3 were obtained (Figure 5F) and could be propagated, while H3K9me1 remained unaffected (Figure S5F). Despite severe depletion of H3K9me2/me3, the interaction between HP1γ and both OCT4 and DPPA4 was retained (Figure 5G), suggesting that the proteins were not interacting exclusively in the context of a H3K9 methyl-containing nucleosome. Thus, from our protein complex isolation, we have identified ESC-specific direct interactors of HP1γ in pluripotent cells.
Differential Interactions and Modifications of the HP1 Proteins in Pluripotent Cells
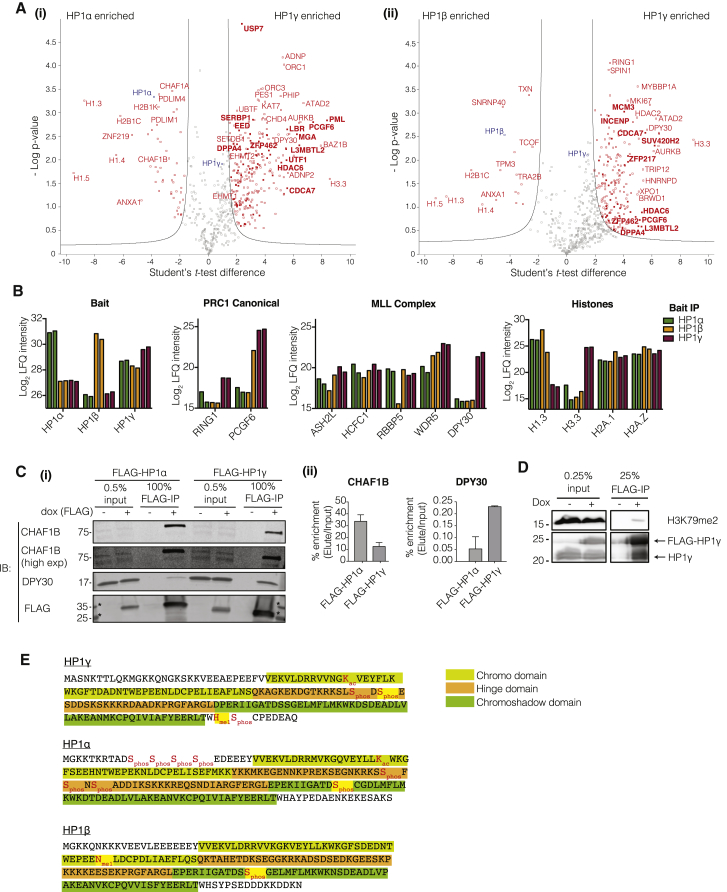
Given the observation that reduction of HP1γ levels increases reprogramming efficiency better than HP1β or HP1α (Sridharan et al., 2013) and the differential localization of the HP1 proteins in ESCs, we next wanted to determine if they participated in distinct complexes. At the protein level, approximately similar numbers of peptides are recovered from each of the HP1 proteins from mouse ESCs, suggesting similar protein expression levels (Graumann et al., 2008, Van Hoof et al., 2006). We generated independent ESC lines that were induced for FLAG-HP1α or FLAG-HP1β expression and subjected the nuclear extracts to IP-MS following the same procedure as for HP1γ and compared the protein profiles obtained (Figure 6A and Table S2). The HP1α-containing complexes were more enriched for several zinc-finger-containing proteins, including ZNF219, a known transcriptional repressor with roles in osteogenic differentiation (Takigawa et al., 2010); the LIM-domain-containing proteins, PDLIM1 and PDLIM4; and the chromatin assembly factor (CAF1) component CHAF1B (Figure 6A). In HP1β there was greater enrichment of TCOF (Figure 6A), a transcription factor involved in Treacher-Collins syndrome that affects neural crest development (Valdez et al., 2004), which may explain the neuronal defects in the HP1β knockout mice.
Figure 6.
Differential Protein Enrichment between HP1α, HP1β, and HP1γ
(A) Volcano plots showing quantitative comparison of proteins enriched in (i) HP1γ versus HP1α, or (ii) HP1γ versus HP1β in ESC extracted with MCN + 0.3 M NaCl. Significantly enriched proteins, red; bait, blue; significantly enriched proteins that are also expressed >5-fold higher in ESC, bold red.
(B) Bar graphs of differential enrichment of proteins between HP1α, HP1β, and HP1γ within known complexes. Data from two independent immunoprecipitation experiments for each group are presented.
(C) (i) IP with FLAG antibody in FLAG-HP1α or FLAG-HP1γ ESC with immunoblot for CHAF1B, DPY30, and FLAG. (ii) Quantitation of percentage enrichment (elute/input) for CHAF1B and DPY30 in FLAG-HP1α or FLAG-HP1γ ESC. Error bars represent SDs of two independent immunoprecipitation experiments.
(D) Western blot of co-immunoprecipitation of H3K79me2 with FLAG-HP1γ.
(E) Post-translational modifications (PTMs) of HP1α, HP1β, and HP1γ found in ESC in red. Previously unidentified PTMs are in red with yellow highlights. Chromodomain, chromoshadow domain, and hinge domain are indicated.
When comparing the bait proteins themselves, HP1α and HP1β were better enriched in their own complexes compared with HP1γ, which was present at similar levels in HP1α and HP1β complexes (Figure 6B). This may suggest a tendency for HP1α and HP1β to homodimerize and for HP1γ to heterodimerize in ESCs. DPY30, which is a core subunit of the MLL-H3K4 methyltransferase complex, was more enriched in the HP1γ pull-down (Figure 6B). Similarly, RING1, a component of both the E2F6 and PRC1-like complexes, and the components GATAD2B and HDAC2 of the NuRD complex, were more abundant in the HP1γ interactions (Figure 6B). Members of the CAF-1 complex are more enriched with HP1α pull-down (Figure 6A). We confirmed the increased association of DPY30 with HP1γ and CHAF1B with HP1α by performing FLAG immunoprecipitation followed by western blotting (Figure 6C). These data suggest that these specific components could partition the chromatin-modifying complexes between the different HP1 proteins.
Interestingly, there was a striking enrichment for histone H3.3 in the HP1γ pull-down, while variants of H2A, such as H2A.Z, remained constant in abundance between the HP1 pull-downs (Figure 6B). In contrast, both the HP1α and HP1β were enriched for histone H1. Given the differential association of histone H1 and H3.3, the HP1α interactome in ESCs resembles the HP1γ interactome in pre-iPSCs (Figure 3B).
We probed these histone associations further by querying the histone post-translational modifications that were detectable. Both HP1γ and HP1β were associated with H3K79me2, a transcription elongation mark (Jonkers and Lis, 2015), and HP1γ with H3K23ac, which is associated with active chromatin (Lu et al., 2015) (Figure S6A and Table S3). We confirmed the association of HP1γ with H3K79me2 by immunoblotting for this modification after IP with FLAG-HP1γ (Figure 6D). In addition to known histone modifications on histone H3 and H4 associated with HP1α and HP1β in somatic cells (Leroy et al., 2012), we found H2BK5 acetylation and H1S1 acetylation, both histone modifications of unknown function (Figure S6A and Table S3). Note that the mass spectrometry analysis was not tailored to detect histone modifications on the N-terminal tail and hence H3K9 methylation was not identified in this analysis.
The HP1 proteins themselves are decorated with multiple post-translational modifications (Leroy et al., 2009, Lomberk et al., 2006a, Lomberk et al., 2006b) in somatic cells, but the status of these modifications in pluripotent cells is unknown. We found a previously unreported phosphorylation on S97 of the hinge domain of HP1γ (Figures 6E and S6B and Table S3) in ESCs. Both S132 and S129 phosphorylations were detected on HP1β and HP1α, but not in HP1γ (Figure 6E). These modification positions are part of the chromoshadow domain, suggesting that some of the HP1 protein functions could differ because of the different post-translational modifications (Figure 6E and Table S3).
Discussion
Pluripotent cells have smaller blocks of heterochromatin as imaged by microscopy (Ahmed et al., 2010, Fussner et al., 2011). The histone H1 and HP1 proteins have lower residence time in chromatin and therefore are much more mobile in pluripotent cells compared with differentiated cells (Melcer and Meshorer, 2010, Meshorer et al., 2006, Meshorer and Misteli, 2006). In this study, we have found that HP1γ, which becomes almost completely nucleoplasmic in mouse ESCs, increases in its association with histone H3.3 but decreases with histone H1 compared with pre-iPSCs. H3.3 is a replication-independent histone that is deposited during transcription elongation. Our earlier results using chromatin immunoprecipitation sequencing (ChIP-seq) in pre-iPSCs and in ESCs have shown that the distribution of HP1γ in pre-iPSCs is both at the promoter and in the coding region of the genes, while in ESCs the binding is largely restricted to the gene body (Sridharan et al., 2013). Taken together these results imply that HP1γ becoming more nucleoplasmic may aid in its recruitment to transcriptionally active regions.
The change in reprogramming efficiency with temporal depletion of HP1γ suggests that in somatic cells HP1γ is required for responsiveness to the reprogramming factors. Later in reprogramming, after HP1γ is in a nucleoplasmic configuration, the transient depletion of HP1γ may allow release from histone H1 association and lead to the gain of pluripotency.
HP1γ interacts with several pluripotency proteins, including OCT4 and ESRRB. One intriguing interaction is with DPPA4, a SAP-domain-containing protein of unknown function that is exclusively expressed in ESCs. Previous studies had suggested a lack of interaction between HP1α and DPPA4 due to the non-overlapping nucleoplasmic distribution and an overlap of DPPA4 with RNA polymerase by immunofluorescence (Masaki et al., 2007). The association of DPPA4 and HP1γ may suggest a different role for DPPA4 in transcription elongation or splicing. While OCT4 is associated with transcription activation and polycomb-mediated repression, from our study it is intriguing that OCT4 also associates with HP1γ. A role for OCT4 in splicing has not been reported, suggesting that OCT4 and DPPA4 may interact with HP1γ in independent complexes.
The HP1 proteins are expressed in many tissues and participate in several chromatin-altering multi-subunit complexes. The preferential binding of HP1γ to some components of these complexes, such as DPY30 of the MLL complex and GATAD2B of the NuRD complex, suggests that the function of the complexes may change depending on the type of HP1 bound. The affinity of recombinant HP1 proteins to H3K9 methylation is affected by post-translation modification of HP1(Hiragami-Hamada et al., 2011). It is interesting to note that the post-translational modifications on HP1α and HP1β were not detected in HP1γ, although they occur in the highly conserved chromoshadow domain. These results suggest that post-translational modifications may regulate the ability of HP1 proteins to participate in specific chromatin-modifying complexes in pluripotent cells. Taken together, our results provide evidence for the compartmentalization of the HP1 proteins in both localization and participation in complexes as cells transition in and out of pluripotency (Figure 7). Although the core pluripotency transcriptional network is conserved between mouse ESCs and human ESCs, they differ in several aspects, including dependence on distinct signaling networks for maintaining pluripotent characteristics. It will be intriguing to determine if the specific interactions and putative functional compartmentalization of the HP1 proteins is conserved in human pluripotent cells.
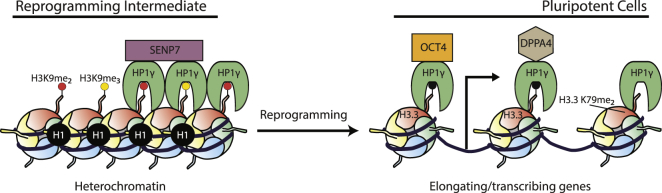
Figure 7.
Proposed Model of HP1γ Interactions
In non-pluripotent cells and reprogramming intermediates, HP1γ interacts with SENP7, which anchors HP1γ to H3K9me2/3 histones in the heterochromatin, depicted as red or yellow circles. HP1γ is also more enriched with linker histone H1 in reprogramming intermediates. During reprogramming, the release of HP1γ from being anchored to the heterochromatin is required to attain pluripotency. In pluripotent cells, HP1γ interacts with histone H3.3 and histone elongation mark H3K79me2. Moreover, OCT4 and DPPA4 are among the interactors of HP1γ in pluripotent cells that occur independently of H3K9me3 but may be dependent on other modifications (black circle).
Experimental Procedures
Derivation and Maintenance of Cell Lines
N-terminal 3X-FLAG-tagged HP1α, HP1β, or HP1γ were recombined into the tet-inducible V6.5 ESC line (Beard et al., 2006). FLAG-tagged protein expression was induced with 1 μg/mL doxycycline. FLAG-HP1γ ESC cell lines with V5-tagged H3.3 wild-type (WT), V5-tagged H3.3 K9M mutant, V5-tagged Dppa4 WT, V5-tagged Dppa4 V81S mutant, or V5-empty were generated by lentiviral transduction. Guide RNAs targeting HP1α (GTTGGACAGGCGCATGGTTAAGG) or HP1β (CTTGATCGGCGAGTTGTCAAGG) were used to generate knockout ESC lines as described by Mali et al. (2013). Pre-iPSC line (Sridharan et al., 2013) with FLAG-HP1γ or FLAG-empty single or with V5-tagged H3.3 were generated by lentiviral transduction. All cell lines were maintained on irradiated MEFs in ESC media (KnockOut DMEM, 15% fetal bovine serum (FBS), l-glutamine, penicillin/streptomycin (pen/strep), non-essential amino acids, 2-mercaptoethanol, and leukemia inhibitory factor). MEFs were cultured in DMEM, 10% FBS, and penicillin/streptomycin media.
Reprogramming Experiments
Reprogramming was initiated with 2 μg/mL doxycycline in MEFs homozygous for Oct4-Klf4-Sox2-Myc allele and heterozygous for the rtTA allele, as described (Tran et al., 2015). For live-cell time-lapse imaging, reprogrammable MEFs were lentivirally transduced with N-terminally tagged GFP HP1γ, and selected with Geneticin for 5 days. Pre-iPSC conversion was induced with ascorbic acid (AA, 50 mg/mL; Sigma) and 2i (1 mM PD-0325901; Stemgent; 3 mM CHIR-99021; Stemgent), and siRNA transfection was performed as in Tran et al. (2015). All procedures involving animals were approved by the Institutional Animal Care and Use Committee of University of Wisconsin-Madison.
Protein Extraction and Immunoprecipitation
Cell pellets were extracted in buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, phosphatase inhibitor and protease inhibitor cocktail) followed by nuclear pellet extraction in buffer C (0.42 M KCl, 20 mM HEPES [pH 7.9], 0.2 mM EDTA [pH 8.0], 5% glycerol, 1 mM DTT, phosphatase and protease inhibitor cocktail); or in micrococcal nuclease digestion buffer (MCN; 50 mM Tris [pH 7.6], 1 mM CaCl2, 0.2% Triton X-100, phosphatase inhibitor and protease inhibitor cocktail) with 20 units of S7 micrococcal nuclease per 1 × 105 cells for 10 min at 37°C and stopped with 50 mM EDTA. NaCl to appropriate levels was added and incubated at 4°C for 1 hr before centrifuging at 13,000 g for 30 min to isolate soluble nuclear extracts. Percentage of HP1γ in the supernatant (S) or pellet (P) fraction was determined by quantifying the immunoblot band intensity using ImageJ of each fraction over the total (S + P).
For FLAG-IP-MS, 20mg of nuclear extract was incubated with 150 μL of FLAG-M2 agarose beads (Sigma, 50% slurry) for 2 hr at 4°C, washed thrice with MCN buffer with 0.3 M NaCl, followed by washing twice with the same buffer but without Triton X-100. IP complexes were eluted with 150 ng/μL 3X FLAG peptide (Sigma) in MCN buffer without Triton X-100. Eluted protein complexes were trichloroacetic acid (TCA) precipitated, in-solution digested with trypsin (Wisniewski et al., 2009), and injected on an Orbitrap mass spectrometer (LTQ Velos, Thermo Fisher Scientific). Three independent immunoprecipitations for pre-iPSC FLAG-HP1γ, and two independent immunoprecipitations for ESC FLAG-HP1γ, FLAG-HP1α, and FLAG-HP1β were performed. For immunoblots, 1.5 mg of nuclear extract was immunoprecipitated with 10 μg of specific antibody or immunoglobulin G (IgG) overnight, collected with 100 μL of Protein G magnetic beads (Bio-Rad) and eluted in Laemmli sample buffer.
For endogenous IP with Benzonase, nuclear extracts were dialyzed to remove EDTA using a 7000 MCO Dialysis cassette (Thermo Fisher Scientific) overnight at 4°C, and 167 U of Benzonase (Sigma) was added per milliliter of dialyzed extracts, supplemented with MgCl2 to 1.5 mM and incubated at 4°C for 2 hr before proceeding with immunoprecipitation as above.
Mass Spectrometry Analysis
Thermo RAW files from tandem mass spectrometry (MS/MS) were analyzed using MaxQuant (Cox et al., 2014, Cox and Mann, 2008) version 1.5.6.5 and searched against the reviewed Mus musculus Swiss-Prot proteome database using the following parameters: trypsin/P digest enzyme with strict trypsin specificity; fixed mod, carbamidomethylation; variable mod, oxidation of methionine; fragment ion mass tolerances, 20 ppm; precursor mass tolerances, 4.5 ppm; peptide false discovery rate (FDR), 1%; protein FDR, 1%. To calculate protein intensities across samples, the options “Match between runs”, and label-free quantification (LFQ) were selected to obtain MaxLFQ. The output file “proteinGroup.txt” was further analyzed using Perseus version 1.5.6.0 for downstream statistical analyses of the normalized protein intensities. Protein lists were filtered to remove potential contaminants, hits to the reverse decoy database, and those only identified by modified peptide. We required that each protein be identified in all independent IP-MS samples from each group. Protein LFQ intensities were logarithmized and missing values imputed by values simulating noise around the detection limit. Contaminants from non-nuclear organelles were removed from the final list of proteins by performing DAVID GO analysis (https://david.ncifcrf.gov/). We performed two-sided t tests with permutation-based FDR cutoff of 1% (S0 = 3). Lists of known interactors of HP1 were obtained from BIOGRID, a database of interaction dataset (https://thebiogrid.org/), as well as previous HP1 IP-MS studies in various cell types (Nozawa et al., 2010, Rosnoblet et al., 2011, Vermeulen et al., 2010). Members of complexes were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/gene) and GeneCards (www.genecards.org).
Author Contributions
Conceptualization, R.S. and N.Z.Z.; Investigation, N.Z.Z., K.J.W., J.E.B., and M.S.; Formal Analysis, N.Z.Z., L.V.S., and M.R.S.; Supervision of Live-Cell Imaging, G.I.; Writing, R.S. and N.Z.Z.; Funding Acquisition, R.S. and L.M.S.; Supervision, R.S. and L.M.S.
Acknowledgments
We thank Prof. Peter Lewis for the gift of H3.3 K9M plasmids and Daniel Devine and César Martinez for technical assistance. This work was supported by the Shaw Scientist award and NIH R01GM113033 to R.S. and NIH-NIGMS R01GM114292 to L.M.S. N.Z.Z. was supported by a pre-doctoral fellowship from the UW-Madison Stem Cell and Regenerative Medicine Center and L.V.S. by the NIGMS-T32GM008349.
Published: January 18, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, three tables, and one movie and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.12.016.
Accession Numbers
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PRIDE: PXD006817 and can be accessed at http://www.ebi.ac.uk/pride/archive/projects/PXD006817.
Supplemental Information
(i) List of proteins enriched in FLAG-HP1γ complexes of ESCs extracted with 0.42 M KCl, MCN with 0.15 M NaCl, or MCN with 0.3 M NaCl against each uninduced sample as control. The union of all HP1γ-enriched proteins from each IP are shown and marked if they were expressed higher in ESCs, and if they were known HP1-interactors.
(ii) List of proteins enriched in FLAG-HP1γ complexes from pre-iPSCs compared with control FLAG-empty vector-transduced pre-iPSCs. Lists of proteins enriched in FLAG-HP1γ complexes from ESCs were also shown as comparison, as used in Figure 2B.
(iii) Quantitative comparison of FLAG-HP1γ IP-MS in ESCs versus pre-iPSC as shown in Figure 3B.
(iv) List of complexes with HP1γ-enriched proteins in pre-iPSCs and ESCs, as shown in Figure 3C.
(i) List of proteins enriched in FLAG-HP1γ, FLAG-HP1α, and FLAG-HP1β complexes in ESCs against each uninduced sample as control. The union of all HP1-enriched proteins from each IP are shown and marked if they were expressed higher in ESCs and if they were known HP1-interactors.
(ii) Quantitative comparison of proteins enriched in HP1γ versus HP1α IP-MS in ESCs extracted with MCN + 0.3 M NaCl, as shown in Figure 6A.
(iii) Quantitative comparison of proteins enriched in HP1γ versus HP1β IP-MS in ESCs extracted with MCN + 0.3 M NaCl, as shown in Figure 6A.
(i) List of modified histone peptides identified in Morpheus G-PTM-D search of HP1γ, HP1α, and HP1β IP-MS in ESCs.
(ii) List of modified HP1γ, HP1α, and HP1β peptides identified in Morpheus G-PTM-D search of each IP-MS in ESCs.
Scale bar, 10 μm.
References
- Ahmad K., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Ahmed K., Dehghani H., Rugg-Gunn P., Fussner E., Rossant J., Bazett-Jones D.P. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aucott R., Bullwinkel J., Yu Y., Shi W., Billur M., Brown J.P., Menzel U., Kioussis D., Wang G., Reisert I. HP1b is required for development of the cerebral neocortex and neuromuscular junctions. J. Cell Biol. 2008;183:597–606. doi: 10.1083/jcb.200804041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A., Zegerman P., Partridge J., Miska E., Thomas J., Allshire R. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Brown J.P., Bullwinkel J., Baron-Lühr B., Billur M., Schneider P., Winking H., Singh P.B. HP1gamma function is required for male germ cell survival and spermatogenesis. Epigenetics Chromatin. 2010;3:9. doi: 10.1186/1756-8935-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A., De La Rosa-Velázquez I.A., Ramirez F., Barenboim M., Onishi-Seebacher M., Arand J., Galán C., Winter G.E., Engist B., Gerle B. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 2014;55:277–290. doi: 10.1016/j.molcel.2014.05.029. [DOI] [PubMed] [Google Scholar]
- Caillier M., Thénot S., Tribollet V., Birot A.-M., Samarut J., Mey A. Role of the epigenetic regulator HP1γ in the control of embryonic stem cell properties. PLoS One. 2010;5:e15507. doi: 10.1371/journal.pone.0015507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussner E., Djuric U., Strauss M., Hotta A., Perez-Iratxeta C., Lanner F., Dilworth F.J., Ellis J., Bazett-Jones D.P. Constitutive heterochromatin reorganization during somatic cell reprogramming. EMBO J. 2011;30:1778–1789. doi: 10.1038/emboj.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann J., Hubner N.C., Kim J.B., Ko K., Moser M., Kumar C., Cox J., Scholer H., Mann M. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell Proteomics. 2008;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- Hiragami-Hamada K., Shinmyozu K., Hamada D., Tatsu Y., Uegaki K., Fujiwara S., Nakayama J.-I. N-terminal phosphorylation of HP1 alpha promotes its chromatin binding. Mol. Cell. Biol. 2011;31:1186–1200. doi: 10.1128/MCB.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Su T., Xue Y., Cheng C., Lay F.D., McKee R.A., Li M., Vashisht A., Wohlschlegel J., Novitch B.G. Cbx3 maintains lineage specificity during neural differentiation. Genes Dev. 2017;31:241–246. doi: 10.1101/gad.292169.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard P.H., Grzenda A., Mathison A., Morbeck D.E., Fredrickson J.R., de Assuncao T.M., Christensen T., Salisbury J., Calvo E., Iovanna J. The Aurora A-HP1gamma pathway regulates gene expression and mitosis in cells from the sperm lineage. BMC Dev. Biol. 2015;15:23. doi: 10.1186/s12861-015-0073-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy G., Chepelev I., DiMaggio P.A., Blanco M.A., Zee B.M., Zhao K., Garcia B.A. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol. 2012;13:R68. doi: 10.1186/gb-2012-13-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy G., Weston J.T., Zee B.M., Young N.L., Plazas-Mayorca M.D., Garcia B.A. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol. Cell Proteomics. 2009;8:2432–2442. doi: 10.1074/mcp.M900160-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.W., Muller M.M., Koletsky M.S., Cordero F., Lin S., Banaszynski L.A., Garcia B.A., Muir T.W., Becher O.J., Allis C.D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G., Bensi D., Fernandez-Zapico M.E., Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 2006;8:407–415. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- Lomberk G., Wallrath L., Urrutia R. The heterochromatin protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Chen X., Sanders D., Qian S., Zhong X. High-resolution mapping of H4K16 and H3K23 acetylation reveals conserved and unique distribution patterns in Arabidopsis and rice. Epigenetics. 2015;10:1044–1053. doi: 10.1080/15592294.2015.1104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan B., Madan V., Weber O., Tropel P., Blum C., Kieffer E., Viville S., Fehling H.J. The pluripotency-associated gene Dppa4 is dispensable for embryonic stem cell identity and germ cell development but essential for embryogenesis. Mol. Cell Biol. 2009;29:3186–3203. doi: 10.1128/MCB.01970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Bailly D., Roche D., Montes de Oca R., Probst A.V., Vassias I., Dingli F., Lombard B., Loew D., Quivy J.-P., Almouzni G. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat. Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- Maison C., Romeo K., Bailly D., Dubarry M., Quivy J.-P., Almouzni G. The SUMO protease SENP7 is a critical component to ensure HP1 enrichment at pericentric heterochromatin. Nat. Struct. Mol. Biol. 2012;19:458–460. doi: 10.1038/nsmb.2244. [DOI] [PubMed] [Google Scholar]
- Maksakova I.A., Thompson P.J., Goyal P., Jones S.J., Singh P.B., Karimi M.M., Lorincz M.C. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin. 2013;6:15. doi: 10.1186/1756-8935-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Saldivia J., van den Bergen J., Krouskos M., Gilchrist M., Lee C., Li R., Sinclair A.H., Surani M.A., Western P.S. Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells. 2006;25:19–28. doi: 10.1634/stemcells.2006-0269. [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki H., Nishida T., Kitajima S., Asahina K. Developmental pluripotency-associated 4 (DPPA4) localized in active chromatin inhibits mouse embryonic stem cell differentiation into a primitive ectoderm lineage. J. Biol. Chem. 2007;282:33034–33042. doi: 10.1074/jbc.M703245200. [DOI] [PubMed] [Google Scholar]
- Mattout A., Aaronson Y., Sailaja B.S., Ram E.V.R., Harikumar A., Mallm J.-P., Sim K.H., Nissim-Rafinia M., Supper E., Singh P.B. Heterochromatin protein 1β (HP1β) has distinct functions and distinct nuclear distribution in pluripotent versus differentiated cells. Genome Biol. 2015;16:1–21. doi: 10.1186/s13059-015-0760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E., Gafken P.R., Ahmad K., Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcer S., Meshorer E. Chromatin plasticity in pluripotent cells. Essays Biochem. 2010;48:245–262. doi: 10.1042/bse0480245. [DOI] [PubMed] [Google Scholar]
- Meshorer E., Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Nakagawa M., Ichisaka T., Shiota A., Yamanaka S. Essential roles of ECAT15-2/Dppa2 in functional lung development. Mol. Cell. Biol. 2011;31:4366–4378. doi: 10.1128/MCB.05701-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L., Ortiz J.A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa R.-S., Nagao K., Masuda H.-T., Iwasaki O., Hirota T., Nozaki N., Kimura H., Obuse C. Human POGZ modulates dissociation of HP1. Nat. Cell Biol. 2010;12:719–727. doi: 10.1038/ncb2075. [DOI] [PubMed] [Google Scholar]
- Ohzeki J.-I., Shono N., Otake K., Martins N.M.C., Kugou K., Kimura H., Nagase T., Larionov V., Earnshaw W.C., Masumoto H. KAT7/HBO1/MYST2 regulates CENP-A chromatin assembly by antagonizing Suv39h1-mediated centromere inactivation. Dev. Cell. 2016;37:413–427. doi: 10.1016/j.devcel.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo K., Louault Y., Cantaloube S., Loiodice I., Almouzni G., Quivy J.-P. The SENP7 SUMO-protease presents a module of two HP1 interaction motifs that locks HP1 protein at pericentric heterochromatin. Cell Rep. 2015;10:771–782. doi: 10.1016/j.celrep.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Rosnoblet C., Vandamme J., Völkel P., Angrand P.-O. Analysis of the human HP1 interactome reveals novel binding partners. Biochem. Biophys. Res. Commun. 2011;413:206–211. doi: 10.1016/j.bbrc.2011.08.059. [DOI] [PubMed] [Google Scholar]
- Shchuka V., Malek-Gilani N., Singh G., Langroudi L., Dhaliwal N., Moorthy S., Davidson S., Macpherson N., Mitchell J. Chromatin dynamics in lineage commitment and cellular reprogramming. Genes. 2015;6:641–661. doi: 10.3390/genes6030641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood A., Hon G.C., Jin F., Henry R.E., Espinosa J.M., Ren B. CBX3 regulates efficient RNA processing genome-wide. Genome Res. 2012;22:1426–1436. doi: 10.1101/gr.124818.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers J.F., Henikoff S. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol. 2001;21:2555–2569. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Gonzales-Cope M., Chronis C., Bonora G., McKee R., Huang C., Patel S., Lopez D., Mishra N., Pellegrini M. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat. Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y., Naruse C., Costa Y., Shirakawa T., Tachibana M., Sharif J., Kezuka-Shiotani F., Kakiuchi D., Masumoto H., Shinkai Y.I. HP1γ links histone methylation marks to meiotic synapsis in mice. Development. 2011;138:4207–4217. doi: 10.1242/dev.064444. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takigawa Y., Hata K., Muramatsu S., Amano K., Ono K., Wakabayashi M., Matsuda A., Takada K., Nishimura R., Yoneda T. The transcription factor Znf219 regulates chondrocyte differentiation by assembling a transcription factory with Sox9. J. Cell Sci. 2010;123:3780–3788. doi: 10.1242/jcs.071373. [DOI] [PubMed] [Google Scholar]
- Thiru A., Nietlispach D., Mott H.R., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K.A., Jackson S.A., Olufs Z.P.G., Zaidan N.Z., Leng N., Kendziorski C., Roy S., Sridharan R. Collaborative rewiring of the pluripotency network by chromatin and signalling modulating pathways. Nat. Commun. 2015;6:1–14. doi: 10.1038/ncomms7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez B.C., Henning D., So R.B., Dixon J., Dixon M.J. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. USA. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D., Passier R., Ward-Van Oostwaard D., Pinkse M.W.H., Heck A.J.R., Mummery C.L., Krijgsveld J. A quest for human and mouse embryonic stem cell-specific proteins. Mol. Cell Proteomics. 2006;5:1261–1273. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- Vermeulen M., Eberl H.C., Matarese F., Marks H., Denissov S., Butter F., Lee K.K., Olsen J.V., Hyman A.A., Stunnenberg H.G., Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Vizcaino J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Yearim A., Gelfman S., Shayevitch R., Melcer S., Glaich O., Mallm J.-P., Nissim-Rafinia M., Cohen A.-H.S., Rippe K., Meshorer E., Ast G. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015;10:1122–1134. doi: 10.1016/j.celrep.2015.01.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(i) List of proteins enriched in FLAG-HP1γ complexes of ESCs extracted with 0.42 M KCl, MCN with 0.15 M NaCl, or MCN with 0.3 M NaCl against each uninduced sample as control. The union of all HP1γ-enriched proteins from each IP are shown and marked if they were expressed higher in ESCs, and if they were known HP1-interactors.
(ii) List of proteins enriched in FLAG-HP1γ complexes from pre-iPSCs compared with control FLAG-empty vector-transduced pre-iPSCs. Lists of proteins enriched in FLAG-HP1γ complexes from ESCs were also shown as comparison, as used in Figure 2B.
(iii) Quantitative comparison of FLAG-HP1γ IP-MS in ESCs versus pre-iPSC as shown in Figure 3B.
(iv) List of complexes with HP1γ-enriched proteins in pre-iPSCs and ESCs, as shown in Figure 3C.
(i) List of proteins enriched in FLAG-HP1γ, FLAG-HP1α, and FLAG-HP1β complexes in ESCs against each uninduced sample as control. The union of all HP1-enriched proteins from each IP are shown and marked if they were expressed higher in ESCs and if they were known HP1-interactors.
(ii) Quantitative comparison of proteins enriched in HP1γ versus HP1α IP-MS in ESCs extracted with MCN + 0.3 M NaCl, as shown in Figure 6A.
(iii) Quantitative comparison of proteins enriched in HP1γ versus HP1β IP-MS in ESCs extracted with MCN + 0.3 M NaCl, as shown in Figure 6A.
(i) List of modified histone peptides identified in Morpheus G-PTM-D search of HP1γ, HP1α, and HP1β IP-MS in ESCs.
(ii) List of modified HP1γ, HP1α, and HP1β peptides identified in Morpheus G-PTM-D search of each IP-MS in ESCs.
Scale bar, 10 μm.